An Introduction to Primer Tools
This feature has been recently added to Base-By-Base to allow users to add primer sequence annotations to their DNA sequences. A primer annotation is displayed and handled in a similar way to a comment.
- The primer sequence
- The primer name
- Location (in your fridge/freezer; contains room for two locations)
- Tm of the primer
- Comments (free text; e.g. last primer use, expiration date, type of primer, etc.)
Adding a Primer:
There are two ways to add a primer: manually (one at a time), or from a file containing one or more primers. Both are available from the “Primers” section of the “Tools” menu.
To add a primer manually, use the “Select mouse mode” to select the region of the DNA sequence that the primer matches. Select “Add Primer” from the “Tools” menu and fill in the primer information.
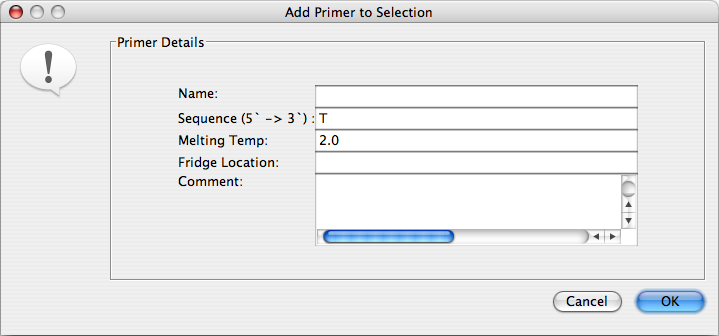
If you are not sure of where the primer is located on the sequence, or wish to add multiple primers, use the “Add Primers from File” option. The file must be a text file containing the following information, separated by tabs: Sequence; Primer name; Location 1; Location 2; Tm; Comments.
A sample primer file follows:
AATCGGGGTACCAAATCT tab Primer-101 tab BoxA1-Freezer1 tab BoxB4-Freezer2 tab 44 tab Write interesting comments here.
AATCGGGGTACAGCTGGA tab Primer-999 tab BoxC1-Freezer1 tab BoxE4-Freezer1 tab null tab General text comments
AATCGCGGGGTACGACTG tab Primer-Joe5 tab BoxG6-Freezer2 tab null tab null tab Some more comments
The program will extract the primer information from the file and will then, for each primer, perform a fuzzy search of the entire sequence for regions that closely match the primer sequence. You will be asked to supply a minimum cutoff point for the % identity desired between primer and sequence. After the search completes, a results screen will display the regions that match the primer sequence. Clicking on the bottom middle button, “Save Primers,” will save all selected primers to the sequence as primer annotations. The bottom right button, “Load Next Primer,” closes the results screen and loads the next primer in the file. The “Abort Search” button exits the primer search entirely.
Tips:
– The primer sequence and primer name are required fields.
– If you do not have values for one or more of the other fields (e.g. Tm), enter null in the location (See example line 2 and 3 above). Do NOT just leave the location blank (this will produce an error message.)
Comment lines can be present in the file; they must start with a # character and are ignored when the primers are extracted from the file. – The file MUST NOT contain any blank lines; these will be treated as wrongly formatted primers and the program will be unable to read the file.
Working with Primers
Each base of a primer is coloured blue (match) or red (mismatch) to show the primer’s similarity/dissimilarity to the sequence it is associated with. Since BBB only displays a single DNA strand at a time, the primers appear dark if they are found on the same strand as is currently displayed, and pale if they are part of the non-displayed strand. If you “mouse-over” a primer, the primer annotation details will be displayed.
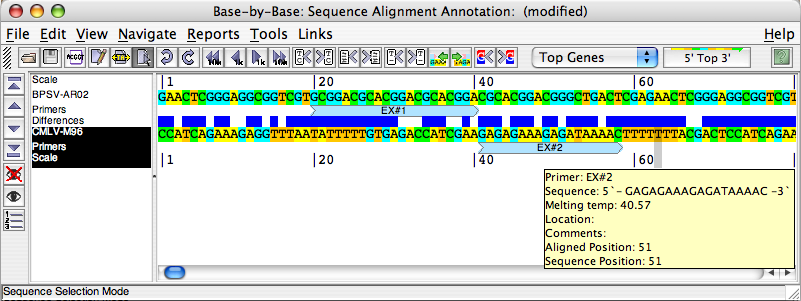
Clicking on the “Last Primer”/”Next Primer” buttons in the toolbar will allow you to navigate through the primers in the sequence (similar to moving from one comment or gap to the next.)
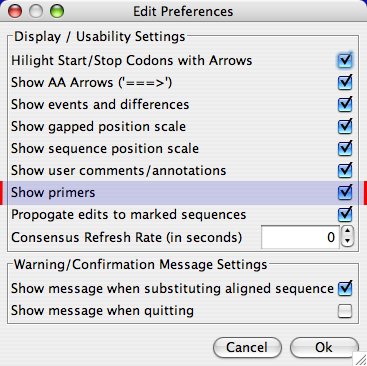
To view primers in the main window, you may need to select the “Show Primers” option from the Preferences panel (File -> Preferences -> Display/Usability Preferences…):
Primer Information Summaries
– All primers associated with a sequence, and their details, can be displayed in a table by selecting “Primer Report” from the “Report” menu. The information displayed can be saved to a file.
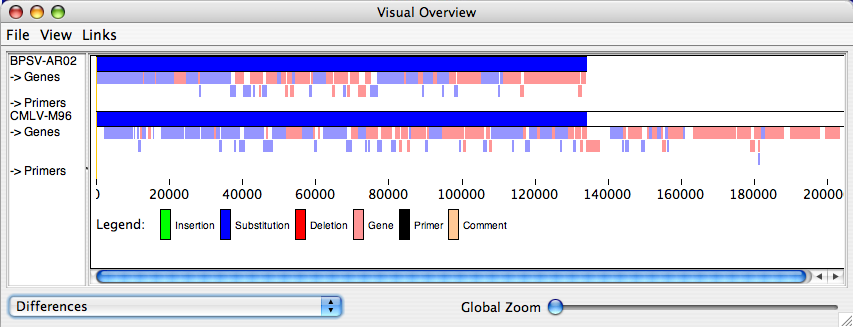
- Primers can be displayed in the Visual Summary, as shown below. Since primers are generally very short compared to the overall sequence, you may wish to focus in on a small area by setting the display area and deselecting the “Show Whole Alignment” option from the Overview window.