Sequence Searcher
NOTE: This tool requires Java Version 8. Download it here: Java Runtime Environment.
With this program, you can:
- search both strands of the nucleotide sequence
- search protein or nucleotide sequences
- search one or more sequences at a time
- search for:
- regular expressions; e.g. GGATCC or TTTTT[ATCG]T
- fuzzy search pattern; e.g. GGATCC allowing one mismatch
- results (all or only a selection of them) can be saved to a file
- choose how to organize your search results (by result, confidence or start/stop).
For a quick How-To for the integrated search just click here.
How the search works
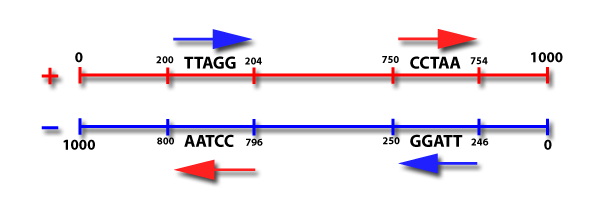
Sequence Searcher looks for every occurrence of the pattern in the target sequences. In the case of nucleic acids, the top strand will be searched in the 5′ -> 3′ direction. Then, the bottom strand will be searched in its 5′ -> 3′ direction for the same pattern. The bases are numbered from 1 at the 5′ end, to the length of the sequence at the 3′ end. See the image below for an example. Searching on an amino acid sequence is straightforward: the sequence is searched for the specified pattern from its start to its stop.
Performance and Limitations
Sequence Length
Sequence Searcher imports sequences approximately at 1 Mbp per second. The program was tested on an older G5 iMac (768MB RAM; 1.6GHz) and found to support sequences for a total of 170 Mbp. This limit is set by the memory of the Java virtual machine and could be increased if required.
Search speed
The following tests were done using a newer Intel iMac (4GB RAM; 2.4 GHz Core 2 Duo).
Fuzzy search
| Search Sequence | Number of mismatches | Time to execute (sec) | Number of hits |
| ACGTACGT | 0 | 2 | 216 |
| ACGTACGTA | 0 | 2 | 44 |
| ACGTACGTA | 1 | 3 | 1808 |
| ACGTACGTA | 2 | 3 | 22124 |
| ACGTACGTACGT | 0 | 1 | 0 |
| ACGTACGTACGT | 1 | 3 | 24 |
| ACGTACGTACGT | 2 | 3 | 672 |
| ACGTACGTACGTACGT | 3 | 4 | 80 |
An exact search (pattern: ACGATCGATC; no mismatches) on five sequences (for a total of 106.48 Mbp) takes approximately 18 seconds. The higher the number of mismatches, the longer the search takes. The same search with 1 mismatch allowed took 83 seconds; with 3 mismatches it took 147 seconds to find 744068 results. Due to the very high number of results, the same Fuzzy search with 4 mismatches used all the Java Virtual Machine memory and could not be completed.
Regular expression search
| Search Sequence | Time to execute (sec) | Number of hits |
| ACGTACGT | 1 | 216 |
| ACGTACGTA | 1 | 44 |
| ACG[TA]ACGTA | 1 | 108 |
| ACG[TA]ACGT[AC] | 1 | 244 |
| ACG[TA]A[CGTA]GT[AC] | 2 | 948 |
| ACG[TA]A.G{1,3}T[AC] | 2 | 1272 |
| ACG[TA]A.{1,50}G{1,3}T[AC] | 3 | 23991 |
Getting Started
If you’re new to Sequence Searcher, click on the launch button (on the right) and use the Quick Start Page to learn the basics (or if you’re like us… just start clicking!).
The VBRC also provides additional help resources for Sequence Searcher:
- How to doc; short descriptions of certain analyses you might want to do.
- FAQs (Frequently Asked Questions)
- Finally, just email us a question and we’ll gladly help you out.
References
If you use this resource please cite the relevant papers (publication list).
Troubleshooting
If your system does not launch Viral Orthologous Clusters, it is probably missing Java Runtime Environment 1.8. Please see Java Web Start Setup and Java Web Start Tips for help.
Submit a feature request
Is there a feature that you think this tool needs? Submit a wish.