Searching in VOCs
There are two ways to search for subsequences in Viral Orthologous Clusters (VOCs). You can search for variations of a subsequence with no mismatches or you can search for variations of a subsequence with a certain number of mismatches. To search for exact matches of a subsequence, you will use “Genomic Sequence Regular Expression Search”. To search for a subsequence with one or more mismatches, you will use “Genomic Sequence Fuzzy Search”. Please note that this page assumes you are somewhat familiar with our VOCs tool already. See the Quick Start Page for VOCs if you need any extra walk-through.
1. Checkmark the box “Select these viruses” (bottom left corner) and select a virus with the sequence that you want to search for:
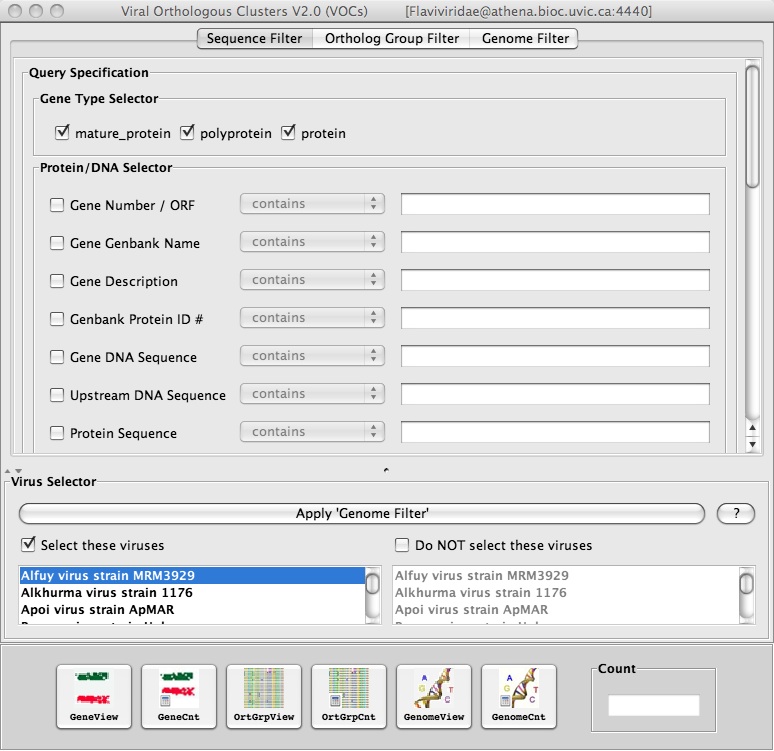
2. To use “Genomic Sequence Regular Expression Search”, click on Search => Genomic Sequence Regular Expression Search and insert a short nucleotide sequence (e.g. GTCATG):
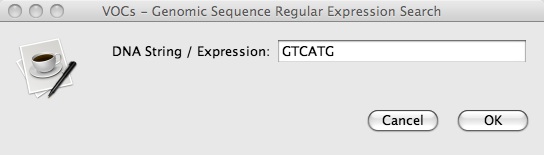
3. Press OK and a new window with a table titled “Genomic Sequence Regular Expression Search” should pop up that looks like this:
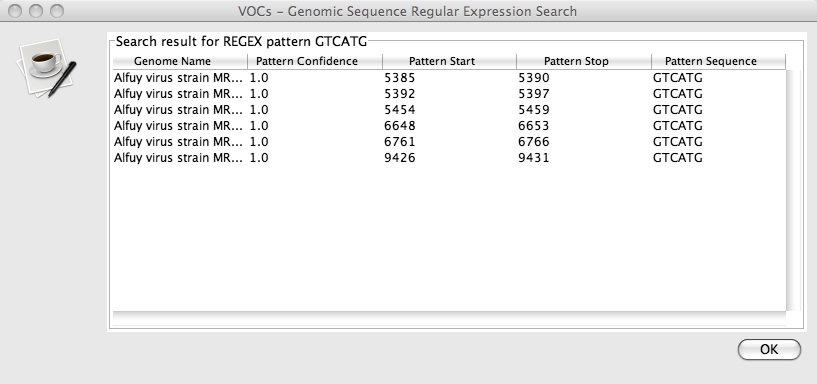
In this case, there are 6 matches with the location of their start and stop positions listed. The pattern confidence indicates the similarity of that subsequence to the one you searched for, which, in this case are all 1.0 because they are exact matches. For more information about the exact nature of the regular expression search click here.
4. To use “Genomic Sequence Fuzzy Search”, click on Search => Genomic Sequence Fuzzy Search, insert a short nucleotide sequence, and set your preference of mismatches (e.g. AGCTAG with 1 mismatch):
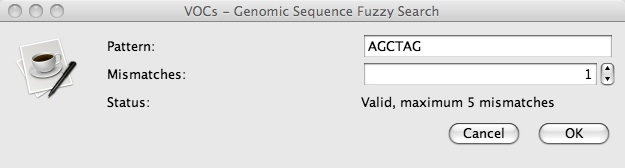
5. Press OK and a new window with a table titled “Genomic Sequence Fuzzy Search” should pop up that looks like this:
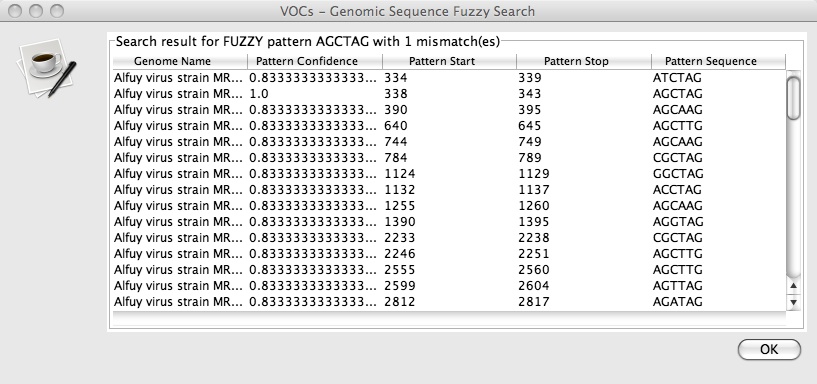
The start and stop positions of each matching sequence are listed along with the position of the mismatch and the pattern confidence. For more information about the exact nature of the fuzzy motif search click here.