Sequence Searcher How-To Page
Note: Though Sequence Searcher acts as a stand-alone program, it is also integrated into the VOCS software. Its functionality and ease of use is much higher within this program. (See details below.)
- Overview of Sequence Searcher
- The interface
- Adding and editing sequences
- Searching the sequences
- Viewing the results
- Saving the results
- Search from VOCS
Overview of Sequence Searcher
Sequence Searcher (SeqS) is an easy to use Java-based tool that allows Fuzzy and Regular Expression searches on multiple sequences. SeqS can perform both search types on DNA and protein sequences.
The Interface
The Menu
The menu bar is very simple and shown below. There are two menus, File and Help. 
File contains the options to save or close the results and to exit the program.
Save Selected Results saves the selected rows of the results table. This option can also be accessed through the keyboard shortcut Alt+S. If there are no results, a warning message is displayed.
Save All Results saves all results (all the rows in the results table). It can also be accessed with Alt+A. If there are no results, a warning message is displayed.
Close Results removes the results from the results tab. The interface is cleaned from all data and is ready to accept the results of a new search. Note that it is not mandatory to close the results before starting a new search: they will be closed automatically after prompting to save them. The keyboard shortcut for this option is Alt+C.
Exit simply quits the program. If there are unsaved results, a prompt is shown. The keyboard shortcut for this option is Alt+X.
The Help menu has links to the Quick Start page of SeqS on this website and to this page.
Input Tab
The input tab is the main interface of the program. As seen below, it has buttons to add sequences to the list (Add manually and Import…) and a list that shows what sequences have been added. Below them, there are the components to customise the search: search type, pattern and mismatches. 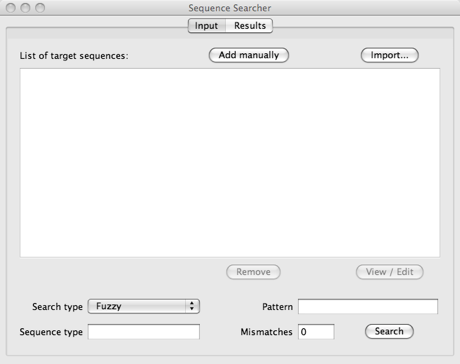
A picture of the application after importing two sequences from the NCBI website and setting some parameters (search type: fuzzy, pattern: acgta, mismatches: 1) is shown below. 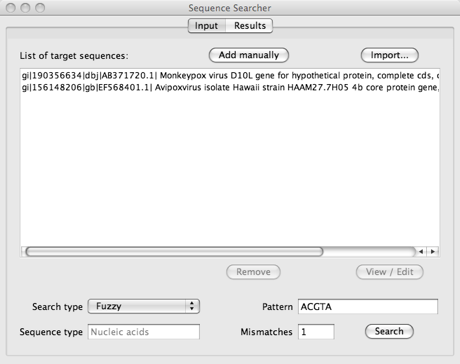
Results Tab
The results tab is automatically selected when a search is complete. Before then it has no data. After performing the search shown above, the results tab looks like this: 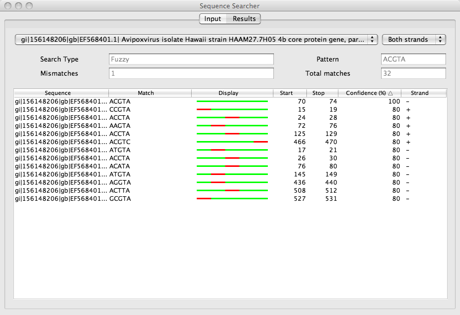
The top left drop down menu is the sequence chooser: it acts as a filter for sequences. With it a user can choose whether to display results from all sequences or from a specific sequence. The drop down menu on the right is a filter for the strands (only used with nucleic acids sequences). After the search parameters you can see the results table. It displays information regarding the matches.
In the case of a Fuzzy search there is a “display” column. It shows where the mismatches (in red) occur within each match. Green marks a perfect match, whereas orange marks a match with a special character (for instance with amino acids B matches both aspartate and asparagine).
On the NCBI website there is a list of ambiguity characters used by Sequence Searcher.
Adding and Editing Sequences
Adding Sequences
Sequence Searcher reads and uses sequences in FASTA format. Two ways of adding target sequences are available: they can be imported from a file or added manually. Please note that all sequences have to be of the same type: nucleic or amino acids, not both.
- Import from file involves choosing a file in FASTA format containing one or more sequences. All sequences, if valid, will be added to the list and labelled by their definition, if specified.
- Manual addition opens a small window where one or more sequences, in FASTA format, can be pasted or typed.
An error message will be displayed if amino acid sequences are present while nucleic acid sequences are being added or vice versa.
Editing Sequences
If there are sequences in the list, it is possible to edit them (one at a time) or remove them (multiple selections allowed) by selecting them and clicking the appropriate button below the list.
- To remove one or more sequences, select them and click the Remove button.
- To edit a sequence, select it and click the View / Edit button: a window pops up and changes can be made. If the change is removing everything from the text area (both definition and description of the sequence), the sequence will be removed.
Searching the Sequences
A search can only be performed if at least one sequence is present in the list, and if a pattern is specified.
The user can select the search type (Fuzzy or Regular Expression) with the search type drop down menu, and the sequences that will be searched.
- If the Fuzzy search is selected, the mismatches field is made available and a number of mismatches can be specified (the valid range is between 0 and the length of the search pattern – 1 inclusive, assuming that a search pattern is not null).
- If the Regular Expression search is selected, only a pattern (a valid regular expression) needs to be specified to perform the search.
See information on Fuzzy and Regular Expression searches.
Once a pattern is specified it is possible to click the search button.
Please note that the search will be performed only on the selected sequences. However, if no sequences are selected, it will be performed on all by default.
Viewing the Results
When the Search button is pressed, the search is performed. When the results are available, the results tab is automatically selected. The target sequences are added to the sequences drop down menu and the data related to the search are displayed.
The table displays the results and they can be sorted by clicking on the column headers.
Filtering results
The drop down menus act as filters: you can filter the results by sequence (all sequences or a particular one) and strand (top, bottom, or both, if applicable).
Saving the Results
The results can be saved to a text file and can be easily imported into spreadsheets such as Microsoft Excel, OpenOffice.org Calc and KOffice KSpread.
Save Selected Results
By choosing this menu option, activated also by the keyboard shortcut Alt+S, only the selected results table rows will be saved to a file. To use it, select the table rows you are interested in saving and choose the menu option Save Selected Results either by clicking on it or by pressing Alt+S.
Save All Results
It is also possible to save all results at once either by clicking the menu option Save All Results or by pressing Alt+A. In this case all results will be saved to a file regardless of the selections on the results table.
Importing Results into a Spreadsheet
Once the results text file has been saved, it is possible to import it into a spreadsheet. To do so open your favourite spreadsheet and open or import the saved text file. The results file format is delimiter-separated values, where the delimiter is a TAB.
Please note that in order to use this functionality the spreadsheet program needs to support the import of delimiter-separated values wp-content/uploads/2018/12.
Search from VOCS
Besides the stand-alone program, Sequence Searcher is also integrated in VOCS. Performing a search in VOCS will call the Sequence Searcher and automatically import the selected sequences.
Search a VOCS database sequence
- Open VOCS.
- For a genome search:
- Select the genome of interest from the list.
- Choose either “Genomic Regular Expression Search” or “Genomic Fuzzy Search” from the Search menu.
- For a gene, protein, or upstream sequence search:
- Find the gene of interest using the search filters/genome list.
- Once the gene has been selected, choose “Search” from the Sequence menu
- Select the relevant search type.
You can find more help on performing sequence searches in VOCS here.