Recent Hits Acquired from BLAST (ReHAB) Documentation
Introduction
ReHAB (Recent Hits Acquired from BLAST) is a tool for finding new protein hits in repeated PSI-BLAST searches. ReHAB compares results from PSI-BLAST searches performed with two versions of a protein sequence database and highlights hits that are present only in the updated database. Results are presented in an easily understandable table, or in a BLAST-like report, using colors to highlight the new hits. ReHAB is designed to handle large numbers of query sequences, such as whole genomes or sets of genomes. Advanced computer skills are not needed to use ReHAB; the graphics interface is simple to use and was designed with the bench biologist in mind.
Sequence similarity searching is a powerful tool to help assign functional, structural, and evolutionary information to DNA and protein sequences. As sequence databases continue to grow exponentially, it is increasingly important to repeat searches at frequent intervals, and these searches tend to retrieve larger and larger sets of results. New and potentially significant results may be buried in a long list of previously obtained sequence hits from past searches.
ReHAB greatly simplifies the problem of evaluating the output of large numbers of protein database searches.
ReHAB Management Console
The components of ReHAB’s main window
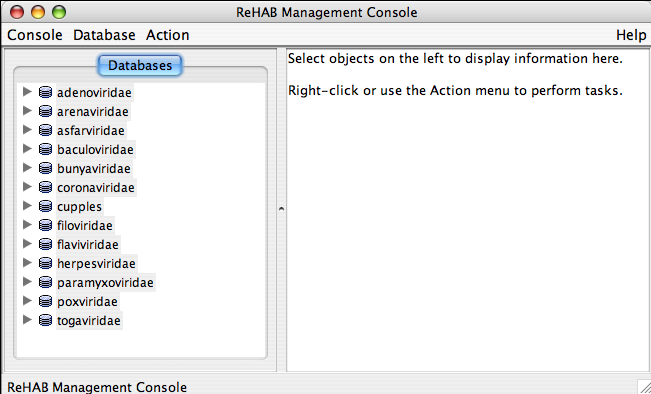
The ReHAB Management Console is the first window that appears in ReHAB. Database information and control is achieved from this window. It consists of 3 main parts:
- The menu bar
- A view listing the databases available
- A view displaying information on a selected database
The Menubar

The Console menu item provides information about ReHAB and the Viral Bioinformatics Resource Center.
The Database menu item allows the user to:
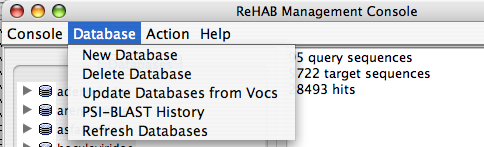
- Refresh the database list.
- We can add new ReHAB database from ReHAB Management console.
- We can delete existing ReHAB database from ReHAB Management console.
- Update Database from VOCs (a VOCs database chooser window appears and viruses are updated from the selected database from here)
- PSI-BLAST History for the selected database (Various dates when the Psi-BLASTs are performed over the DB)
The Action menu item allows one to:
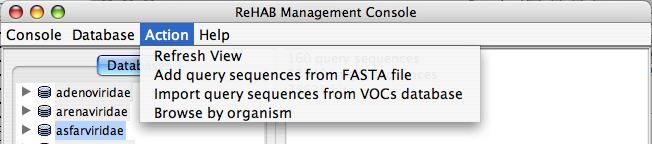
- Refresh the section displaying the information on a selected database
- Browse a selected database
- Add query sequences from FASTA file
- Import query sequences from VOCs database
Databases View
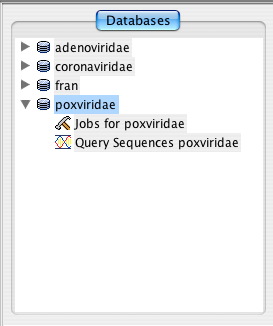
The database view displays a list of the available databases. Within each database listing one can view any long running jobs for that database or view the query sequences submitted for that database.
Information View

The information view provides database specific information such as:
- Statistics
- Long Running Jobs
- Query Sequences
Statistics

Statistical information displayed include:
- The number of query sequences within the chosen database
- The number of target sequences involved in alignments
- The number of significant alignments (hits) between the query and target sequences
Long Running Jobs

The information view can display the status of any long-running PsiBLAST jobs for the chosen database.
Query Sequences
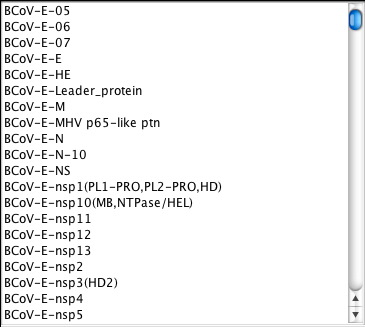
The information view can display the query sequences from the chosen database.
ReHAB Hits Browser Window
The components of ReHAB’s hits browser window
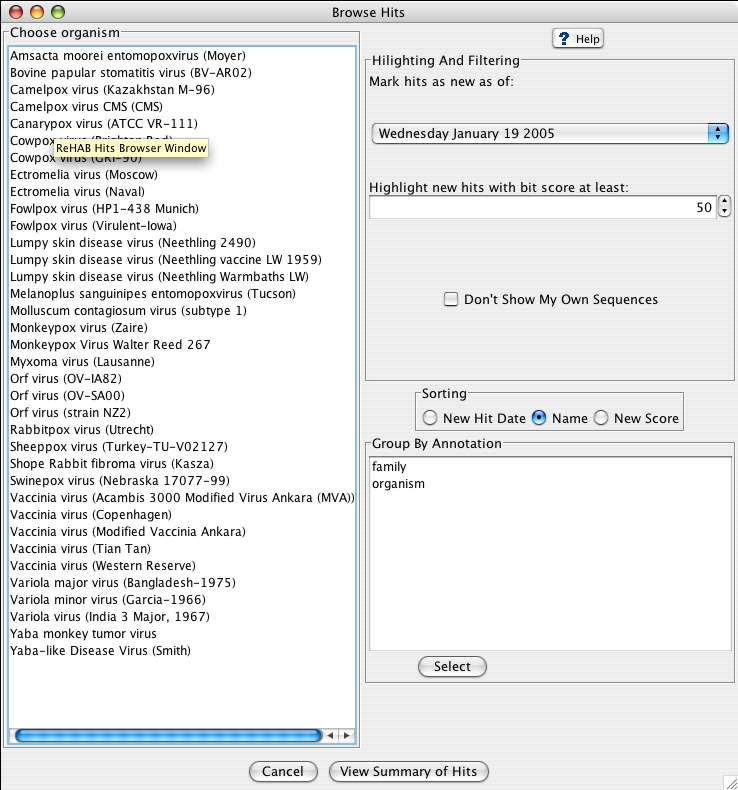
Choosing to browse a database from the ReHAB Management Console results in the Hits Browser Window. This window consists of 4 main parts:
- Choose organism section
- Highlighting And Filtering section
- Sorting section
- Group By Annotation section
Choose Organism

This section displays a list of query sequence sets organized by organism or family annotation. Selecting an item and clicking on View Summary of Hits or double clicking on an item results in another window displaying hit summaries for that query set contingent upon the constraints that the user applied. The various contraints a user can apply are outlined below.
Highlighting And Filtering
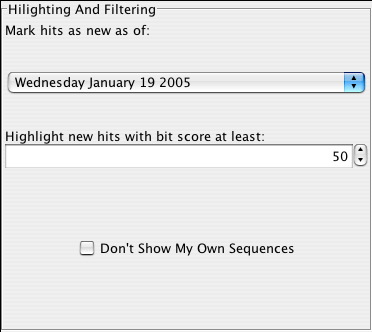
This section allows the user to:
- Mark hits as new as of a specific date on which a search was run. The available dates are those on which the query sequences were searched against a then current nr NCBI database.
- Highlight new hits with bit score at least of a specified value. Since all new hits are not necessarily significant, results are highlighted in different colors depending on the bit score. New hits scoring above the specified threshold are highlighted in red while those scoring below are highlighted in yellow. Since all query sequences that have new hits are highlighted, any that remain unhighlighted do not have new hits.
- Select a checkbox that will filter out identical sequences. Unless a sequence has not been deposited in the public database, a sequence similarity search will return results including the query sequence itself, as well as nearly identical sequences that are orthologs of the query.
Sorting
The output can be sorted based on three criteria (name, new hit date, or maximum new hit bit score) by selecting the appropriate radio button.
Group By Annotation
Selecting family or organism from this section reorganizes the groupings of the query set list based upon the annotation chosen.
ReHAB Summary of New Hits Window
The components of ReHAB’s summary of new hits window
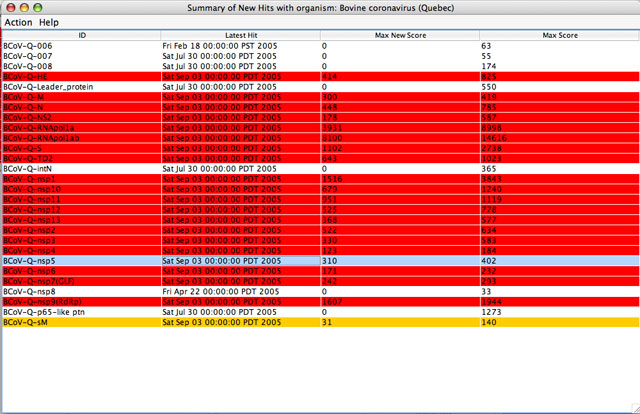
Selecting an item and clicking on View Summary of Hits or double clicking on an item in the Hits Browser Window results in the Summary of New Hits Window displaying hit summaries for that query set contingent upon the constraints that the user applied. This window consists of 2 main parts:
- The menu bar
- A table listing new hit summaries for each query sequence in the query set selected in the Hits Browser Window.
The Menu bar

The Action menu item allows one to view information about the hits in 2 ways:
- Selecting HTML Report launches the user’s default web browser and displays the hit-list for the selected query in familiar BLAST-style.
- Selecting Hits Manager generates a new window that displays the hit-list for the selected query. The Hits Manager Window that is generated allows further analysis of the sequences in the hit-list.
The Help menu item provides help information for the Summary of New Hits Window.
Summary of New Hits Table
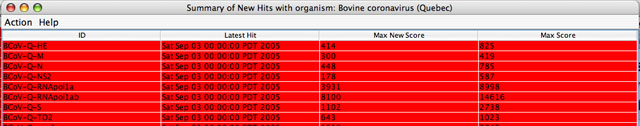
This table consists of 4 column headings:
- ID
- Latest Hit
- Max New Score
- Max Score
Since all new hits are not necessarily significant, results are highlighted in different colors depending on the bit score. The user can change the default threshold of the minimum bit score, to show new hits scoring above this cut-off in red and new hits scoring below it in yellow. Since all query sequences that have new hits are highlighted, any that remain unhighlighted do not have new hits. However, unhighlighted queries may have significant hits from previous searches. TheLatest Hit column indicates this fact: query sequences showing hits only from previous searches show an older hit date and a bit score of “0” in the Max New Score column. Unhighlighted sequences with no information in the Latest Hitcolumn do not have any hits in the database or they been filtered out. Sorting of the entries can be changed by clicking on the column heading. Details about the hits can be obtained by right-clicking on the entry or selecting an option in theAction menu. Max Score column gives the overall maximum score.
ReHAB HTML Report Window
The components of ReHAB’s HTML report window
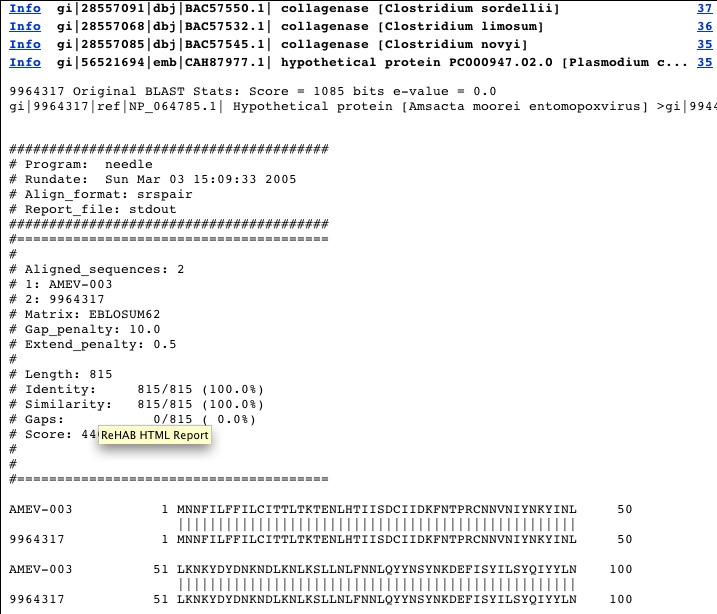
This window is launched from the user’s default web browser and displays the hit-list in familiar BLAST-style. Hits are displayed in descending order of bit score; however, a key feature of this program is that new hits are highlighted in red or yellow. The pairwise alignment can be displayed rapidly by clicking on the score (a hyperlink). In contrast to the usual BLAST output, which presents the local alignment found by BLAST, a global alignment produced by Needle is shown. More information can be obtained about the target sequence by clicking on the link to the NCBI file for that entry.
ReHAB Hits Manager Window
The components of ReHAB’s hits manager window
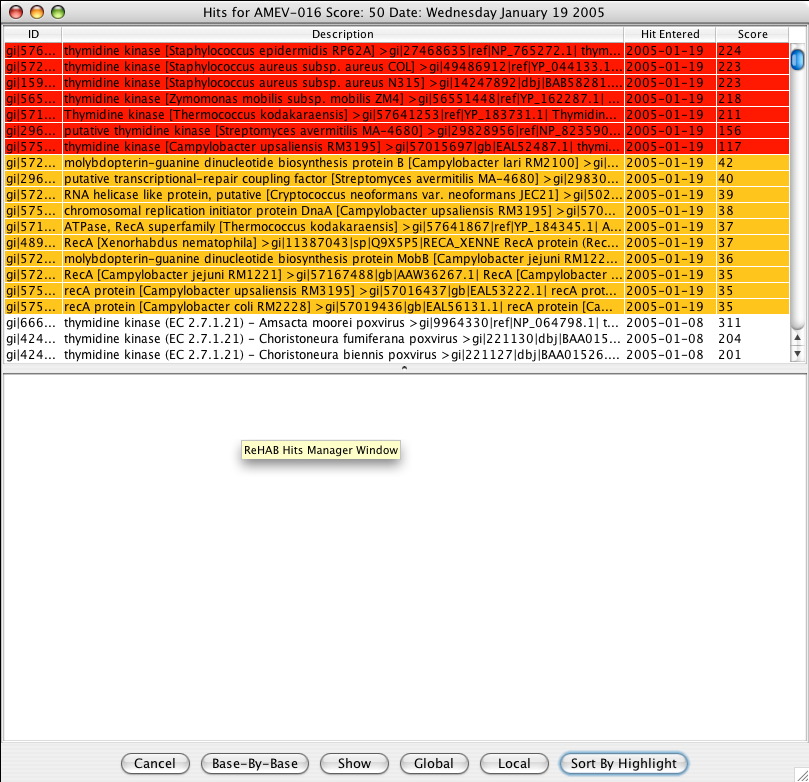
The Hits Manager Window is generated from the Summary of New Hits Window. It consists of 2 main parts:
- A table listing the target sequences hit with the query sequence.
- A workspace displaying various hit analysis output depending upon which button is pressed.
Hits Manager Table
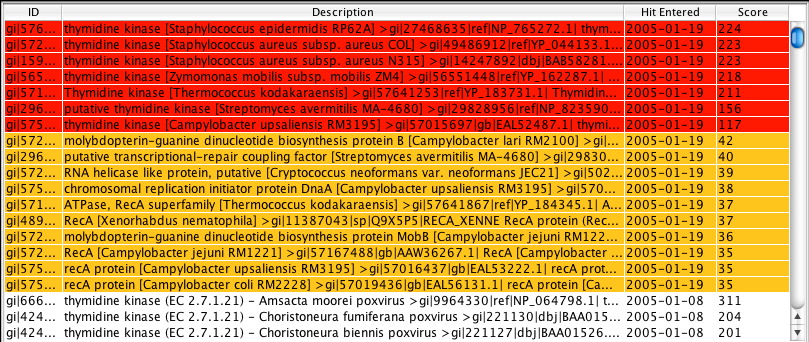
The Hits Manager Table lists the target sequences which hit with the query sequence. The ID column contains the unique gi number for the specific target sequence while the Description column lists more specific textual information regarding the target sequence. The Hit Entered and Score columns provide information as to when the hit was entered in the database and its corresponding bit score with the query sequence, respectively. Table sorting is achieved by clicking on the appropriate column heading. Highlighted sequences in red represent new hits that scored above the bit score threshold while those in yellow represent the new hits that scored below the threshold. Only highlighted sequences represent the new hits that were generated.
Workspace
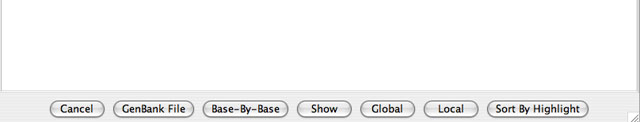
The workspace is the area below the hits manager table. It displays various hit analysis output depending upon which button is pressed. The possible actions and resulting outputs displayed in the workspace are:
- Double clicking a sequence from the hits manager table results in a standard Emboss Needle (Global) alignment output to be displayed in the workspace.
- Clicking Sort By Highlight sorts the hits manager table such that the new (and therefore highlighted) hits are displayed at the top of the table arranged in a descending order based on bit score.
- Clicking on Local after selecting 1 or more target sequences from the hits manager table results in a standard Emboss Water (Local) alignment output to be displayed in the workspace.
- Clicking on Global after selecting 1 or more target sequences from the hits manager table results in a standard Emboss Needle (Global) alignment output to be displayed in the workspace.
- Clicking on Show displays the query sequence in FASTA format followed by any target sequences (also in FASTA format) that were selected from the hits manager table.
- Clicking on Base-By-Base launches the Base-By-Base application already preloaded with the query sequence and any target sequences selected from the hits manager table.
- Clicking on the GenBank File button will take you to the NCBI GenBank file URL corresponding to the genes.
- Clicking on Cancel closes the hits manager window.
ReHAB Tutorials
How to browse a database
Browsing a database in ReHAB can be accomplished in 2 ways:
- Select a database from the database list in the ReHAB Management Console -> From the Action menu select Browse by organism. The Hits Browser window will be generated.
- Double click on the desired database. The Hits Browser window will be generated.
How to view the status of any long running jobs for a database
Viewing the status of any long running jobs for a database requires the ReHAB Management Console. A sideways triangle icon is positioned to the left of each database in the database list. Click on this icon and subitems will appear below the database. Select Jobs for DATABASE_X and the status of any long running jobs for that database will be displayed in the panel to the right.
How to view a database’s list of query sequences
Viewing a database’s list of query sequences requires the ReHAB Management Console. A sideways triangle icon is positioned to the left of each database in the database list. Click on this icon and subitems will appear below the database. Select Query Sequences DATABASE_X and the list of all query sequences submitted for that database will be displayed in the panel to the right.
How to perform alignments
Emboss Needle (Global) and Water (Local) utilities are utilized to perform pairwaise alignments between query and target sequences.
Needle (Global) alignments can be generated in 2 ways:
- From the Summary of New Hits window, select HTML Report (query) from the Action menu item – or – right click a selected query sequence from the table and select HTML Report (query) from the menu that is generated. The default browser will generate an HTML report listing the target sequences followed by their corresponding needle alignments with the query sequence. The hyperlink to the left of each target sequence links to an NCBI page containing information about that specific target sequence. The hyperlink to the right of each target sequence brings the page directly to the corresponding needle alignment of the query sequence with that target sequence.
- From the Hits Manager window, select the desired target sequences from the table and click on the Global button. The output of each pairwise alignment with the query sequence is displayed in the workspace area.
Water (Local) alignments are generated from the Hits Manager window. Select the desired target sequences from the table and click on the Local button. The output of each pairwise alignment with the query sequence is displayed in the workspace area.
How to view the summary of hits for a query set
The Hits Browser window is required to view the summary of hits for a query set.
Firstly, select a query set from the Choose organism list.
In the Highlighting And Filtering section of the window, select a date from the drop down box upon which any new hits will be highlighted. The value in the Highlight new hits with bit score at least field highlights a new hit in red if it is above that threshold value, otherwise it is yellow. Clicking the Don’t show my own sequences checkbox filters out results that are identical or nearly identical orthologs of the query sequence.
The Sorting section of the window sorts the resulting Summary of New Hits window based on the radio button selected (New Hit Date, Name or New Score).
Click View Summary of Hits to generate the Summary of New Hits window for the query set selected based on the constraints applied.
How to input query & target sequences into Base-By-Base for analysis
From the Hits Manager window, select the desired target sequences from the table and click on the Base-By-Base button. The Base-By-Base application will be launched with the query and chosen target sequences pre-loaded for analysis.
How to add and delete ReHAB databases?
We can add a new ReHAB database or delete an exiting ReHAB database from the ReHAB Management console. Select Database menu from the ReHAB Management Console menubar. From there follow New Database or Delete Database. To delete a database ReHAB deletes a selected database from the Database View.
How to Update Query sequences in ReHAB database from VOCs?
From ReHAB Management Console select Database menu, then follow Update Databases from VOCs. A VOCs database chooser window will appear and after we choose a VOCs Database, ReHAB updates the ReHAB database with new query sequences from selected VOCs database.