The Sequence Filter
This is where you may define the type of gene you wish to include in the search; a pseudogene (select “pseudogene“) and/or functional genes known to encode a protein (select “protein“).

If a check mark appears in the box next to the gene type, that feature is selected. To deselect a type of gene, click on the box so that the check mark disappears.
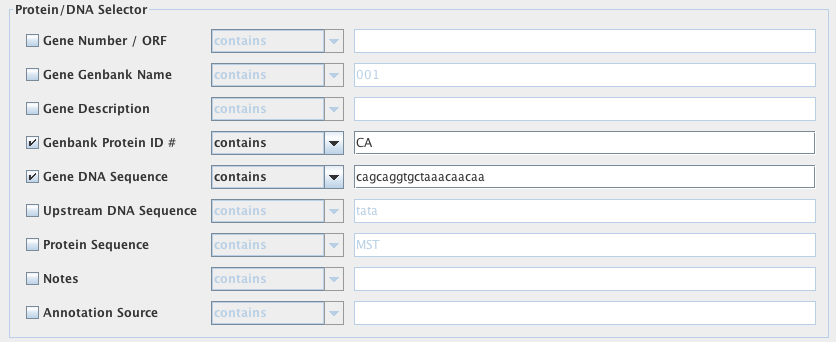
Protein/DNA Selector
Here, you may define constraints on the genes of interest and proteins they encode. In these blank fields, the constraints can be written in either plain text or regular expressions. For each constraint, you may select to have VOCs search for a gene set that “contains“, “equals“, “starts with“, “ends with” or “does NOT contain” the information you decide to input. These options are selected from the drop down menu next to each of the search fields.
Molecular Weight/Isoelectric point/AA Count
If you would like to search for a broader range of genes, you can use this to search for genes encoding proteins that fall within a certain range of physical properties. You may enter a range for the molecular weight, isoelectric point and/or the number of amino acids in the protein. This is particularly useful if you do not know the exact value of the protein’s physical property or if you would like to investigate proteins with properties similar (but not necessarily identical) to the properties of another protein of interest.
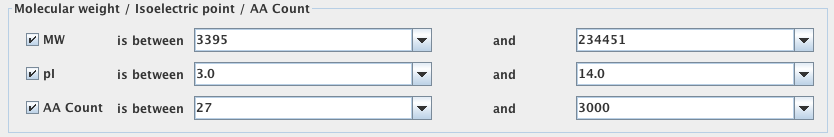
There are drop down menus from which you may select values, however, values may also be entered manually.
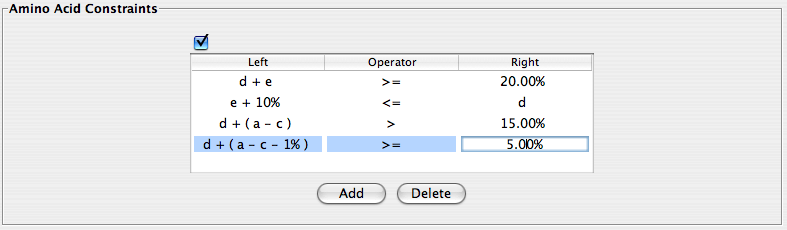
Amino Acid Constraints
If you know at least a small amount of information about the protein sequence you are looking for, this section could prove to be of particular use. This section defines constraints applied to the % composition of amino acids of the protein; the constraints consist of a left parameter, a right parameter, and a comparison operator ( >, <, =, … ). The left and right parameters are entered as simple mathematical expressions containing the one letter codes for the amino acids along with numbers (representing percentages) and various symbols: (+ , – , ( , ) ) and spaces. To enter a criterion, press “Add” right below the window and manipulate the parameters within the appropriate fields. In a similar fashion, a criterion may be deleted by selecting “Delete“.
The IUPAC accepted one letter amino acid codes can be entered in upper or lower case and are as follows:
| Standard Amino Acids | Non-Standard Amino Acids | ||||
|---|---|---|---|---|---|
| AMINO ACID | CODE | AMINO ACID | CODE | AMINO ACID | CODE |
| Alanine | A | Leucine | L | Asparagine or Aspartate | B |
| Arginine | R | Lysine | K | Glutamine or Glutamate | Z |
| Asparagine | N | Methionine | M | Isoleucine or Leucine | J |
| Aspartate | D | Phenylalanine | F | Selenocysteine | U |
| Cysteine | C | Proline | P | Unknown | X |
| Glutamate | E | Serine | S | ||
| Glutamine | Q | Threonine | T | ||
| Glycine | G | Tryptophan | W | ||
| Histidine | H | Tyrosine | Y | ||
| Isoleucine | I | Valine | V | ||
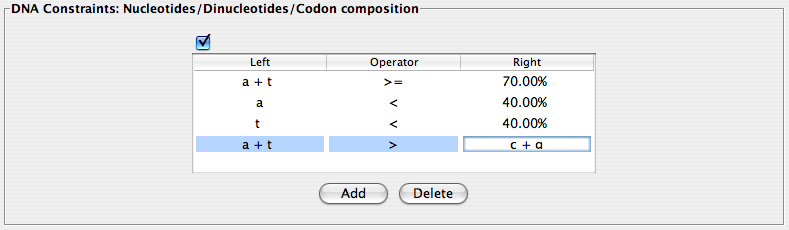
DNA Constraints: Nucleotides/Dinucleotides/Codon composition
In a similar way to the amino acid constraints, the DNA constraints can be entered as a simple mathematical equation using the same symbols and comparison operators described in the amino acid constraints section. Again, VOCs will search for % composition of nucleotides of the gene sequence. Criteria are added to and deleted from the corresponding window in the same way as the amino acid constraints described above.
The IUPAC accepted one letter nucleotide codes can be entered in upper or lower case and are as follows:
| Nucleotide(s) | Code |
|---|---|
| Adenine | A |
| Cytosine | C |
| Guanine | G |
| Thymine | T |
| Unknown base | X |
| Purine (A or G) | R |
| Pyrimidine (C or T) | Y |
| Strongly bonding (G or C) | S |
| Weakly bonding (A or T) | W |
| Keto (G or T) | K |
| Amino (A or C) | M |
| Every base except A (C, G or T) | B |
| Every base except C (A, G or T) | D |
| Every base except G (A, C or T) | H |
| Every base except T (A, C or G) | V |
| Any base (A, C, G or T) | N |