Purpose
The PDB scan, implemented in
VOCs, is designed as a starting point for researchers interested in finding matches between virus sequences and known structures in the PDB database.
Details
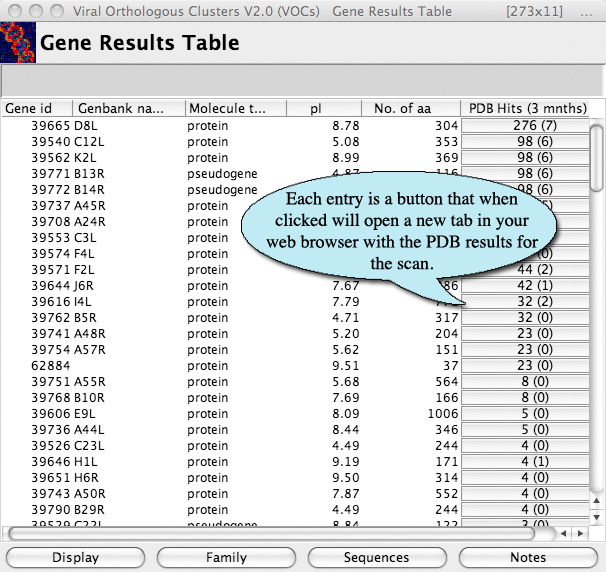
The PDB scans run weekly Thursdays and the results are counted and displayed in the gene viewer in VOCs. The results of the scan are displayed in a button (see figure 1) that when pressed will open the PDB hit for that scan (see figure 2). Each PDB button contains a number and then a number in brackets. The number in brackets indicates the number of hits that are 3 months old or less. This allows for easy scanning of new hits. If there is an asterix after the number, this indicates that the scan is currently running and retrieving the latest numbers.
Figure 1. The number in parentheses represents only the most recent hits in the last three months.
Figure 2. The results of the query will be opened.
For the scan we use blastp with an e-value of .00001 to search the PDB using a reference gene (either from a hand-picked reference genome or simply the gene with the longest sequence) from that homolog group. Because of using the reference gene, the numbers on the PDB button may not be the same as what you would get on the website, but they should be within a close margin. Also affecting our count are PBD’s repeat ids. They use chaining and can count multiple hits with one id, in our version each id is counted only once.
For a summary of the PDB hits you see, click Structure Hits by Date in the PDB menu as seen below in figure 3.
Figure 3. The PDB menu.
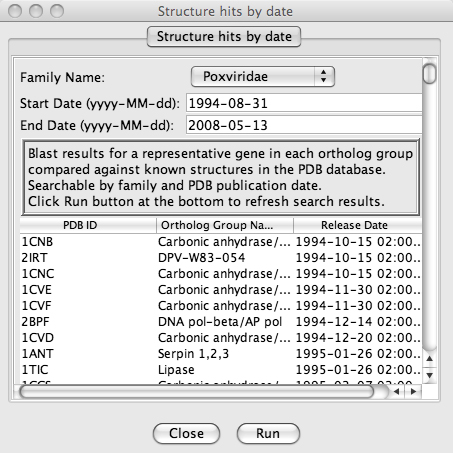
The window in figure 4 (below) sumarizes the hit aquired by the blastp. The results may be limited to certain dates and arranged by date, PDB id or ortholog. Clicking on any entry will open the PDB page with the relevant results.
Figure 4. Summary of the blasts run.