JDotter Quick Start
What is it? JDotter is our Java-based program for creating dot plots for two or more sequences.
What can I do with it? A lot!! Compare large genomes: view repeats/insertions/translocations; compare proteins: overview of conserved regions; analyze suboptimal alignments.
How do I get started? JDotter can be accessed for free at www.virology.ca. From the VBRC Tools drop-down on the left side of the page, select JDotter, then click the Launch Program button. JDotter will be downloaded to your computer and Java Web Start will automatically start the program.
JDotter can also be accessed from within the VOCs database.
What next? The main JDotter window, shown below, will open. From here you can load VOCs or user-supplied sequences into the program and create dot plots.
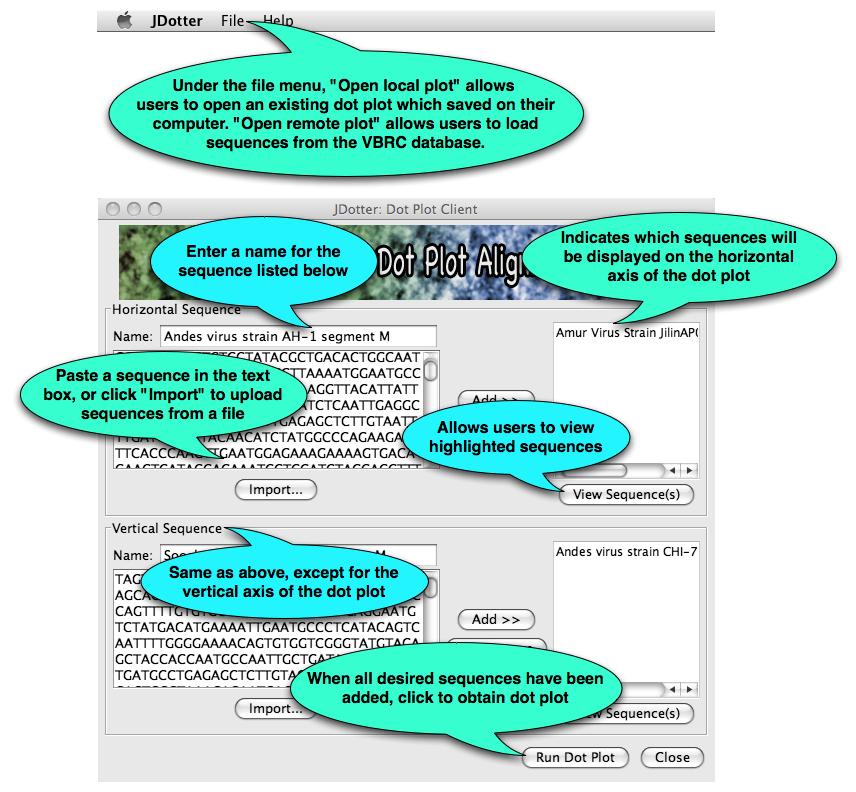
A new window will open with a dot plot of the sequences:

Examples: Here are some of the tasks JDotter can be used for:
- create whole-genome, sub-genome or protein dot plots from user-supplied sequences;
- quickly load pre-calculated plots of complete genomes from VOCs database;
- view and navigate the plots to pinpoint insertions/deletions and evaluate overall genome similarity;
- print/save dot plots in JDotter or JPEG format.