VOCs Quick Start Page
What is it? VOCs (Viral Orthologous Clusters) is our Graphical User Interface for accessing and searching the VBRC viral databases, containing data from 12 virus families.
What can I do with it? VOCs can be used to perform a large variety of tasks, including:
- Search the VBRC databases at the genome, gene, protein, or ortholog-cluster level
- View and save sequence and statistical data in tabular or text format
- Select data and link directly to analysis tools (VGO, BBB, JDotter, etc.)
How do I get started?
- Select VOCs from the VBRC Tools drop-down menu (top left of page).
- Click the Launch Program button. VOCs will be downloaded to your computer and Java Web Start will automatically start the program.
- Choose the database of interest from the list that appears.
Click here to go directly to the VOCs page.
What next? The main VOCs window shown below will open. From here you can search the VOCs database that you selected and open various whole-genome analysis tools.
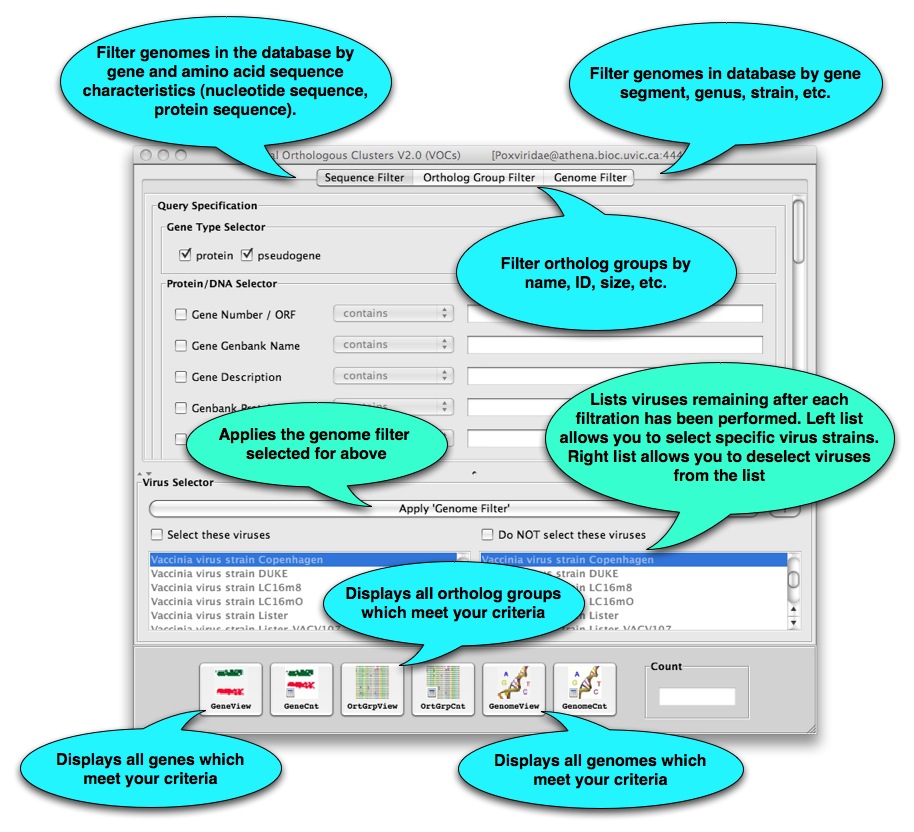
After you select gene, ortholog group, or genome view, a new window will open with an interactive table of your search results:

Examples: Here are some examples of tasks that VOCs can be used for (and the time required).
- select all orthopoxvirus DNA ligases; align proteins (displayed in BBB; 44 sequences): 75 seconds
- select 2 coronavirus genomes; recall or perform whole-genome dotplot; display in JDotter: 30 seconds
- select 10 coronavirus spike proteins; generate pairwise dotplots (100 dotplots total); display in single JDotter window: 45 seconds
- generate gene map of any genome in the database: under 10 seconds
- compare the nucleotide content of five poxvirus genomes using DNAGrapher: 1 minute
- select all poxvirus DNA polymerase genes; export a single FASTA file of all DNA sequences: 45 seconds