eGATU
eGATU
Overview of eGATU
eGATU (easy Genome Annotation Transfer Utility) is a light application modelled on GATU. It allows users to transfer annotations to an input genome without having to specify a reference.
To use eGATU, import a FASTA or GenBank file describing a genome. Select the family it belongs to, and genus if known. Click Run to try to annotate the input genome. If all coding sequences are found, eGATU proceeds to transfer the annotations. Otherwise, input from the user is required in order to select the genes to annotate. The output file, a genome in the GenBank format, can be saved and viewed as text, or opened in VGO together with its reference, or aligned with Base By Base.
The Interface
Menu
The menu bar contains two menus, File and Help.
File contains options for most eGATU functions.
Preferences allows the user to set the BLAST preferences.
Save annotations saves the annotations on the input genome to a text file.
View annotations shows the annotations that have been transferred.
VGO analysis opens both reference and input genome in VGO.
BBB alignment opens the two sequences in Base By Base and aligns them.
Help links to this help page.
Main Window
The interface allows the user to select to what family and genus the genome to annotate belongs, import this genome from a FASTA or GenBank file and annotate it.
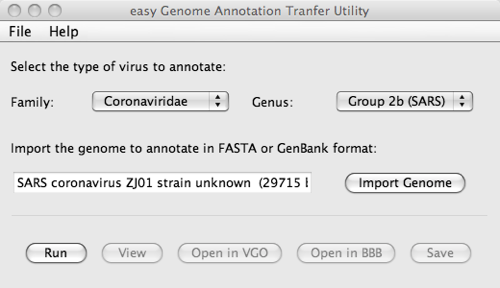
On completion of the process, it is possible to read and save the annotations as well as analyse the reference and target genomes with VGO and Base By Base.
Raw Annotations Window
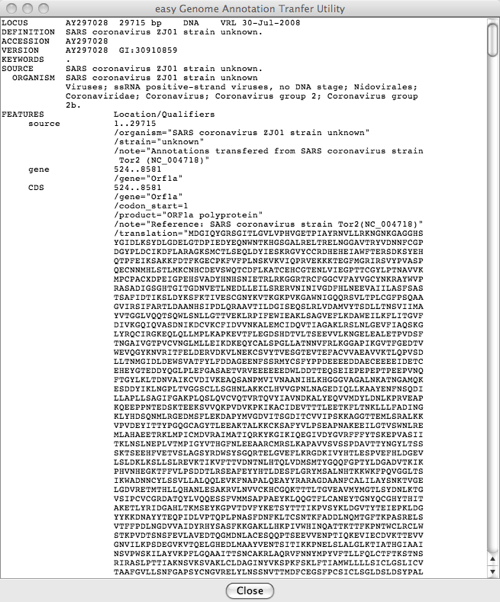
Shows the output (the annotations that have been transferred to the target gnome) in GenBank format. This file is what would be saved with the Save Annotations option.
Gene Table
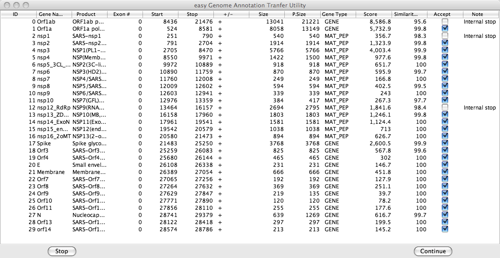
In case not all annotations can be automatically transferred, a gene table is displayed. The table shows information on all genes, such as gene name, product, exon number, start and stop, strand, the length of the gene on the genome to be annotated, the length of the gene on the reference genome, the gene type, the score, the percentage of similarity, whether or not to accept (transfer) it and, in case of an error, a message. User input is now required to proceed to the next stage or to interrupt the process. A screenshot of the gene table is shown below.
The Reference Genome
The target genome is BLASTed against a subset of reference genomes from our database. eGATU automatically chooses a reference, and displays the name and the BLAST score it obtained. If a reference cannot be found, it is recommended to use GATU.
Analyses
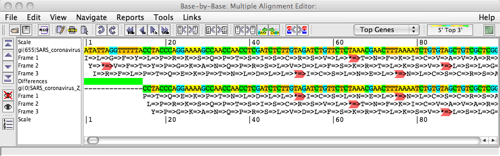
eGATU only annotates a target genome. It allows the user however to analyse it with other tools, such as VGO and Base By Base. An example of the alignment with Base By Base is shown below.