A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes : Nature Communications : Nature Publishing Group
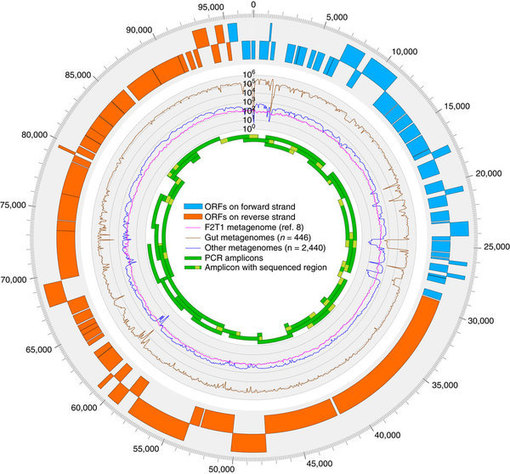
Metagenomics, or sequencing of the genetic material from a complete microbial community, is a promising tool to discover novel microbes and viruses. Viral metagenomes typically contain many unknown sequences. Here we describe the discovery of a previously unidentified bacteriophage present in the majority of published human faecal metagenomes, which we refer to as crAssphage. Its ~97 kbp genome is six times more abundant in publicly available metagenomes than all other known phages together; it comprises up to 90% and 22% of all reads in virus-like particle (VLP)-derived metagenomes and total community metagenomes, respectively; and it totals 1.68% of all human faecal metagenomic sequencing reads in the public databases. The majority of crAssphage-encoded proteins match no known sequences in the database, which is why it was not detected before. Using a new co-occurrence profiling approach, we predict a Bacteroides host for this phage, consistent with Bacteroides-related protein homologues and a unique carbohydrate-binding domain encoded in the phage genome.
Source: www.nature.com
Not quite about Aquatic Viruses, but too cool not to share. PS – love the name!
See on Scoop.it – Aquatic Viruses