Java GUI for InterProScan (JIPS) Documentation
Introduction
Background: Recent, rapid growth in the quantity of available genomic data has generated many protein sequences that are not yet biochemically classified. Thus, the prediction of biochemical function based on structural motifs is an important task in post-genomic analysis. The InterPro databases are a major resource for protein function information. For optimal results, these databases should be searched at regular intervals, since they are frequently updated.
Results: We describe here a new program JIPS (Java GUI for InterProScan), a tool for tracking and viewing results obtained from repeated InterProScan searches. JIPS stores matches obtained from InterProScan searches performed with multiple versions of the InterPro database and highlights hits that have been added since the last search of the database. Results are displayed in an easy-to-use tabular format. JIPS also contains tools to assist with ortholog-based comparative studies of protein signatures.
Conclusions: JIPS is an efficient tool for performing repeated InterProScans on large batches of protein sequences, tracking and viewing search results, and mining the collected data.
JIPS Management Console
Once the JIPS database is selected from the database chooser, JIPS Management Console window appears.
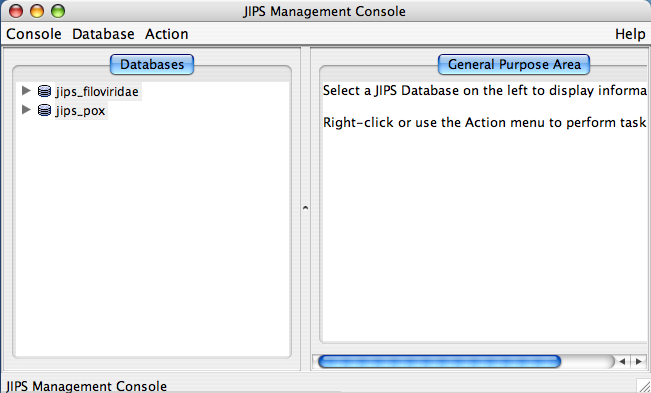
The JIPS Management Console consists of three main parts:
- Menu bar.
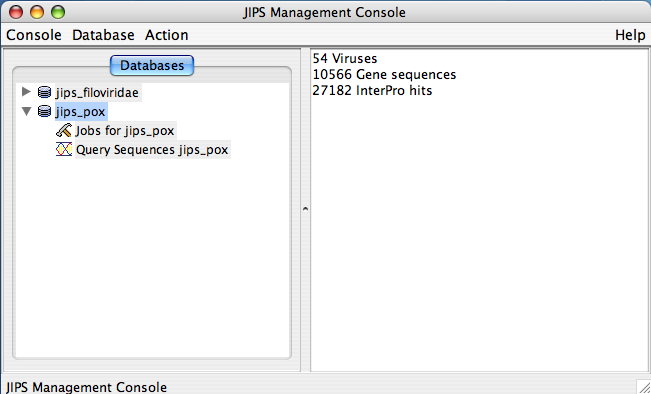
- Database Pane: This is left side window of JIPS Management console, and displays all the available JIPS databases within the JIPS database server. Each JIPS database is represented as a single node. JIPS database node is expanded by clicking on the tringle icon preceeding the database icon for the JIPS database node. Every JIPS database node has two sub nodes a Jobs for JIPS_DB_NAME and a Query sequences for JIPS_DB_NAME.
- Statistics/Status Pane: This is located right side of the main JIPS console. This component displays various information depending upon the node chosen in the Database Pane. If JIPS Database node is selected it displays the statistics for the selected JIPS database. If the Jobs for JIPS_Database is selected then a graphical Progress bar is displayed which tells the current status of the jobs completed. If Query sequences for JIPS_Database is selected then all the query sequence names corresponding to the JIPS database are displayed.
Menu bar
The Menu bar consists of three menus: Console, Database, and Action.
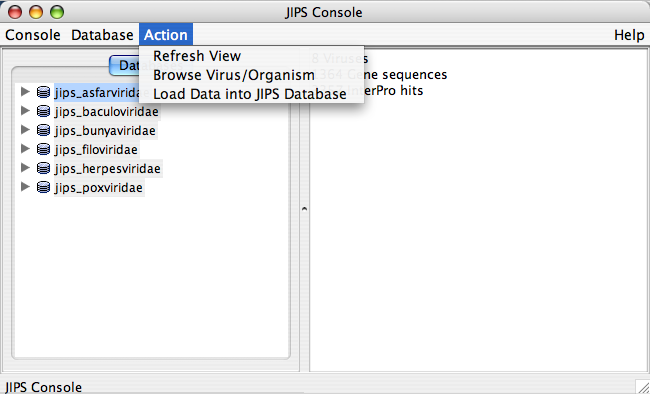
Action Menu
The user needs to select the database before performing any action. Usually Action menu is empty if the database is not selected. Alternatively, the user can select a JIPS database and right click to see all the permitted action events.
Each of the nodes has its own set of actions. So, actions that can be performed on a JIPS database node is different from Jobs fo JIPS_database node. By selecting a JIPS database node or any of its sub nodes, and choosing Actionmenu from the Menu Bar will display permissible actions. The actions allowed also depends on the type of user. Administrative User can perform all actions, where as a Browser user (one who does not have any administrative privileges) can perform only a subset of allowed actions. The following are the actions that can be performed, depending on the type of user (administrator user or browser user):
JIPS Database Node Actions
- Create new JIPS database (Admin user only): Creates a new JIPS database
- Delete existing JIPS database (Admin user only): Deletes selected JIPS database
- Load data into JIPS database (Admin user only): Searches for new sequences from VOCS and updates JIPS local database
- Interproscan History: gives you various dates when InterProScan was performed on this JIPS DB
- Reset Database Window: Refreshes the database list. Usually performed after a Database is created or deleted or updated
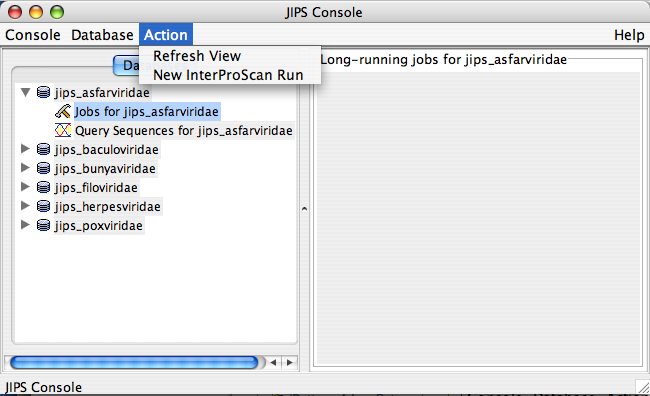
Jobs for JIPS_database Node Actions
This is a sub node of JIPS database node. * Initiate new InterProScans over the JIPS database (Admin user only)
- Status of the InterProScan. This is a progress bar that appears on the JIPS-Statistics/Status Pane. This tells how far the InterProScan are completed for the selected JIPS database.
Query Sequences for JIPS_database Node Actions
This is a sub node of JIPS database node. To delete query sequences or add Family ID to query sequences admin user must select the query sequences of interest. The actions are not enabled unless the query sequences are choosen.
- Refresh view (refreshes the statistics/status pane and displays updated information)
- Delete Query sequences (Admin user only). Deletes selected query sequences from the Statistics/Status Pane
- Add Family ID to the query sequences (Admin user only). This action adds/modifies family ID to the query sequences. Family ID is a NUMBER. This action is generally performed when sequences are loaded from a FASTA file. After the proteins are loaded, groups of protein can be selected from Statistics/Status pane and family ID added to them.
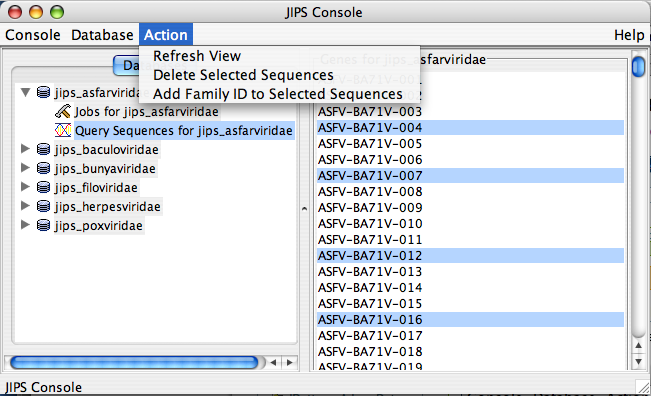
NOTE ALTERNATIVELY THE ACTIONS CAN BE PERFORMED BY 1. RIGHT CLICKING ON THE NODE, WHICH POPS UP A ACTION MENU 2. CHOOSE THE ACTION FROM THE POPUP MENU
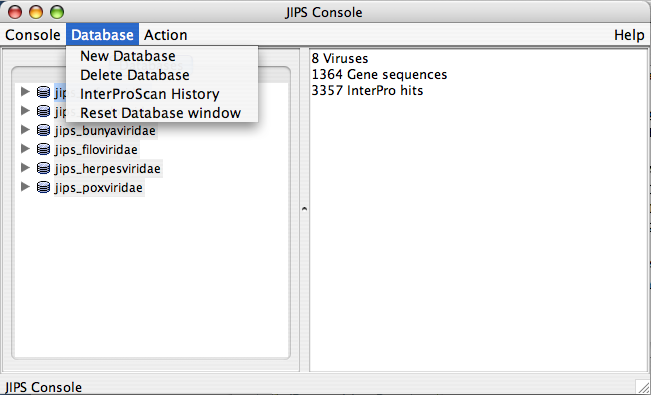
Database Menu
- Refresh view: Refreshes the Database list
- Browse Viruses: Displays all the viruses/organisms from a JIPS database
- Import sequences from VOCs database: Fill JIPS database from VOCs database
- Import sequences from Fasta File: Fill selected JIPS database from a FASTA file.
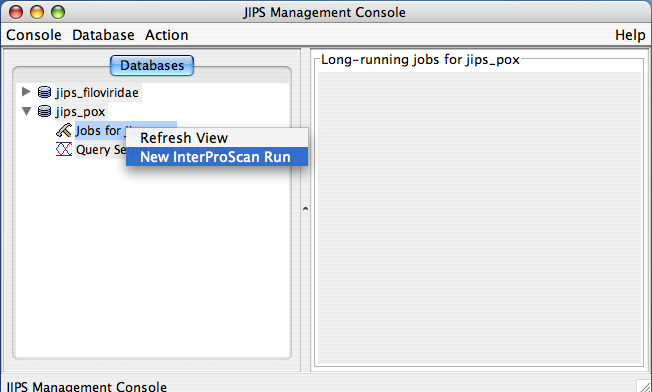
Initiating New InterProScan and Checking Status of submitted job
JIPS user can submit InterProScan runs for every gene with ease. All it requires is couple of mouse clicks. Please note that only Admin user is given permissions to perform this action. Every JIPS database that is present in the JIPS database pane, has two child nodes: Jobs for the JIPS database and Query sequences for JIPS database.
To perform new InterProScan on the JIPS database, the JIPS Admin simply needs to right click on the “Jobs for the JIPS database” and select Run InterProScan Job menu item from the popup. When you just select the “Jobs for the JIPS database” node, JIPS displays current status (a progress bar appears) of initiated InterProScan in the “Statistics/Status Pane” which is the right pane of the JIPS main console.
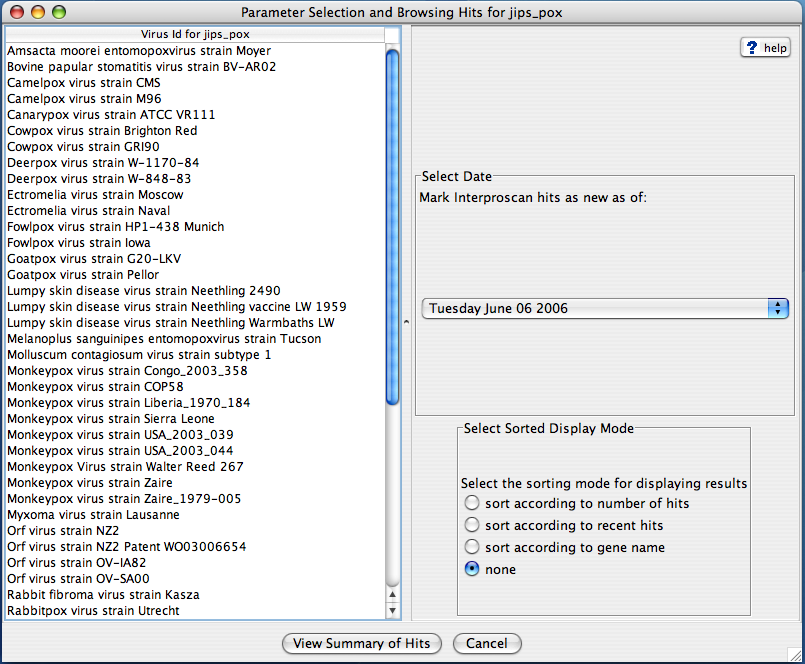
JIPS Browse Virus/Organism
User can browse and select a virus/organism and choose to display the summary of hits. Summary of hits can be displayed according to a cutt off date, and can be sorted according to a few choices (sort according to gene name, number of new hits, total number of hits).
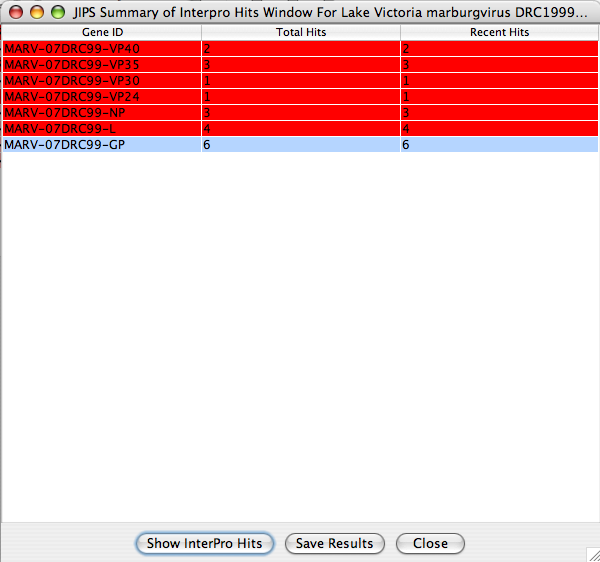
JIPS Summary of Hits
This window displays summary of interpro hits for each gene within the selected virus/organism, which is selected from JIPS virus/organism broswer .
JIPS users can double click any of the displayed genes, and a JIPS Hits Frame will be displayed. Alternatively, users can select the gene and click on Show InterPro Hits button. Users can save this hit information by clicking on Save Resultsbutton. All the hits will be saved into a tab delimited file.
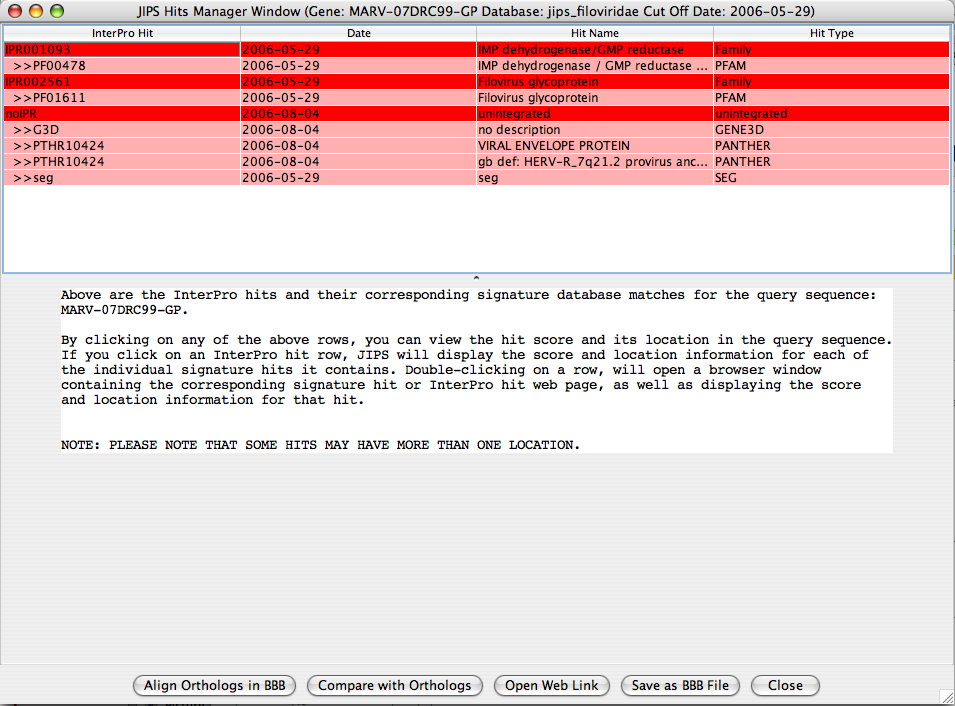
JIPS Hits Frame
JIPS Hits frame contains all the Interpro hits for the selected gene from the JIPS Summary of Hits window. The users can navigate through each hit and visualize which region of the sequence is acctually corresponds to the InterPro/Signature database hit.
By selecting a row in the Singrature databse hit, hit’s score and location information is displayed in the bottom protion of the JIPS Hit Frame window. If the user selects a row which is an InterPro hit, then JIPS displays all the signature database hits that are retrieved under the corresponding InterPro id and for this gene. Protein Sequence panel displays the protein sequence, and the Score information Panel displays the score information of hit(s). When the location button for a pirticular signature database hit is clicked, the amino acids corresponding to this signature hits are colored RED.
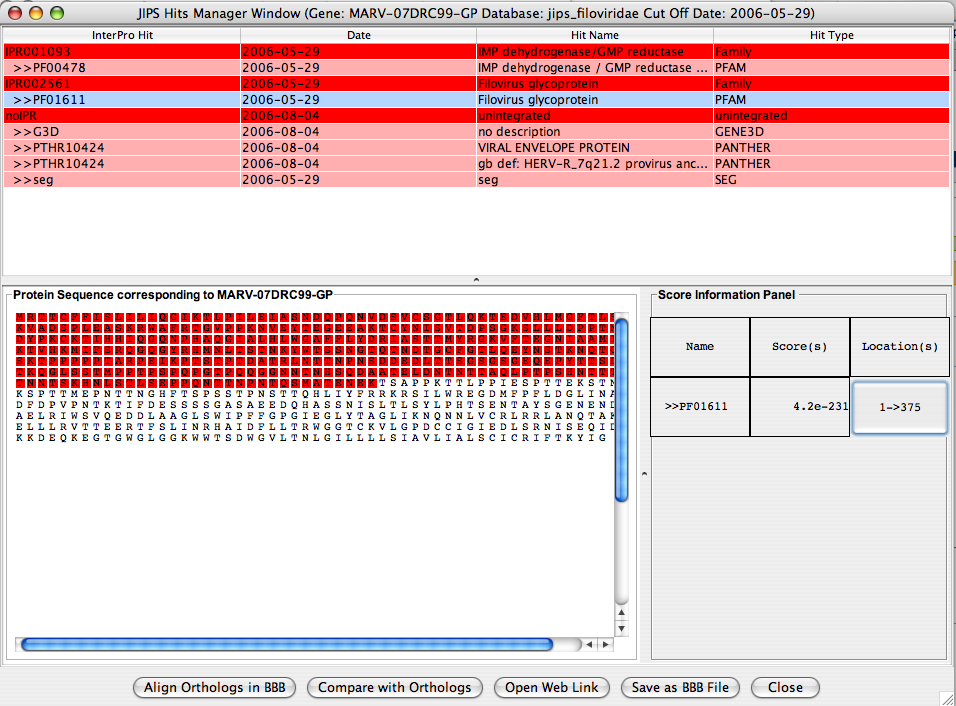
Action Panel
- Align Orthologs in Base-By-Base: When this button is clicked Gene Select Panel (discussed below) is displayed. User can select the Orthologs from a Gene Select Panel . All the selected orthologs will be aligned in the BBB editor along with the query gene sequence and its interpro annotations.
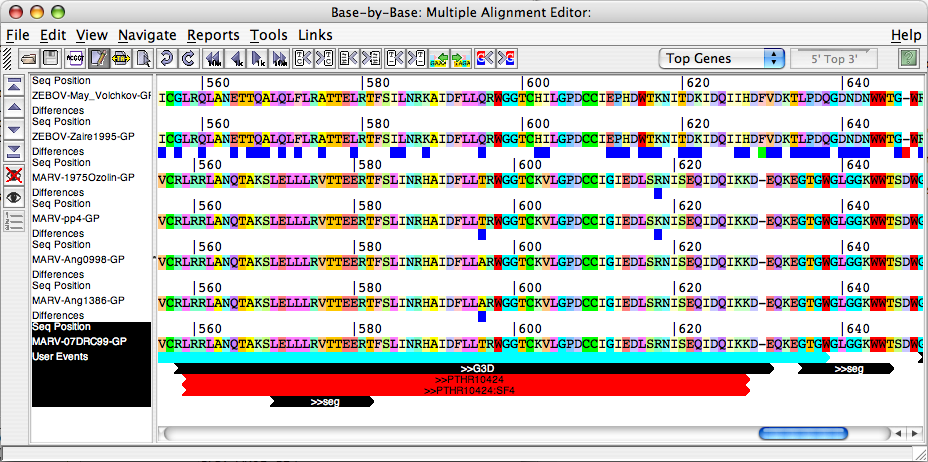
- Compare Orthologs: When this button is clicked Gene Select Panel (discussed below) is displayed. User can select the Orthologs from a Gene Select Panel. JIPS compares Interpro hits that are obtained for the query gene/protein (whose results are displayed in the JIPS Hits Frame) with the Orthologs that are selected from the Gene Select Panel and displays them in a nice graphical window. This window succint information about the conserved signature hits among query and its orthologs.
- Open WWW Link: Opens the weblink for the selected Interpro/signature database with the corresponding database entry. To perform this action user needs to select the hit row.
- Save as BBB: Saves the query sequence as a Base-By-Base file. All the Interpro/signaure hits information is stored as annotations (Comments) of the protein sequence.
Ortholog Select Panel
When the user wants to compare orthologs or align orthologs in Base-By-Base , the user has to select orthologs of his/her interest. This is facilitated through Ortholog Select Panel. User can select multiple Orthologs at once and proceed.
JIPS Orthologs Frame
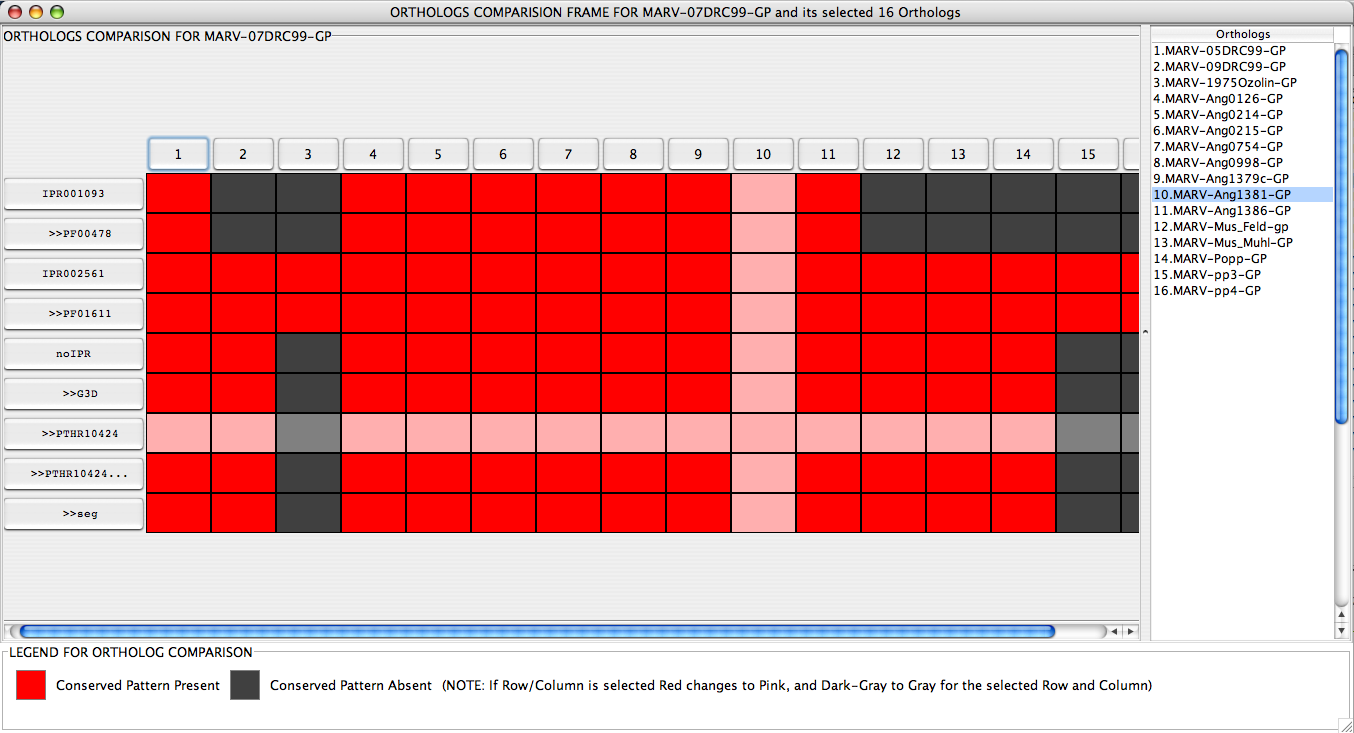
Ortholog Comparison Frame displays the easily comprehendible graphical view of comparison or InterPro hits between the query sequence and its Orthologs. This frame is obtained by selecting Compare with Orthologs button from theJIPS Hits Frame window of a query sequence. As soon as this button is pressed, a Ortholog Selection Panel (This is discussed in the JIPS Hits Frame ) is poped up. User can select Orthologs from this panel and click ok. JIPS retrieves InterPro hit information for each of the selected orthologs and displays in a nice graphical window (see below). All the selected Orthologs are displayed on the X-axis, and are numbered starting from 1. The actual Ortholog name is separately present in a table on the right hand corner of the Ortholog Comparison Frame. A row of the table is automatically selected when a corresponding column in the graphical comparison window is selected, and vice versa. All the Interpro/signature hits for the query sequence are displayed in the first column. Every cell beneath each Ortholog is colored RED or GRAY, representing the presence or absence of the query’s Interpro/Signature databsae hit in the Ortholog’s hits. If a row or column are selected these colors turn to PINK and LIGHT_GRAY.
User can double click Ortholog Button or the table entry to display the JIPS Hits Frame for that ortholog in a separate window. Also, users can double click query’s InterProScan hit buttons and JIPS will open a browser pointing to the database entry.
JIPS Ortholog Comparison Frame
JIPS Tutorials
Content
How to Browse a Database
Browsing a database in JIPS can be accomplished in 2 ways
- Select a database from the database list in the JIPS Console -> From the Action menu select Browse by organism. The Hits Browser window will be generated
- Double click on the desired database. The Hits Browser window will be generated.
How to view a database’s list of query sequences
Viewing a database’s list of query sequences requires the JIPS Console. A sideways triangle icon is positioned to the left of each database in the database list. Click on this icon and subitems will appear below the database. Select Query Sequences under any of the jips_X database and the list of all query sequences submitted for that database will be displayed in the panel to the right. If you are a admin user you may proceed to delete or modify the sequences.
How to view the summary of hits for a query set
The Hits Browser window is required to view the JIPS Summary of Hits Window for a query set.
- Select a query set from the Choose organism list from
JIPS Virus/Organism Broswer
- .
- In the Highlighting And Filtering section of the window, select a
DATE
- from the drop down box upon which any new hits will be highlighted.
- The Sorting section of the window sorts the resulting Summary of New Hits window based on the radio button selected (New Hit Date, Name or New Score).
- Click View Summary of Hits to generate the Summary of New Hits window for the query set selected based on the constraints applied.
How to add and delete JIPS databases
To DELETE or ADD NEW database, user should be a JIPS Administrative user. We can add a new jips database or delete an exiting jips database from the JIPS console. Select Database menu from the JIPS Console menubar. From there follow New Database or Delete Database. To delete a database, user need to select a database, and JIPS deletes a selected database permenantly.
JIPS Requirements
To run JIPS web based clients users need to have Java Webstart (JWS), and J2SE 1.4 or higher are required. JWS usually comes with the JRE.
If users are interested in having there own JIPS server and database server, they require MySQL 4.0 or higher, Interpro databases and Interproscan, and J2SE 1.4 or higher are required.
NOTE JIPS requirse following open source jar wp-content/uploads/2018/12 that are not developed by the VBRC: 1. biojava.1.4.jar,bytecode-0.92.jar we obtained a copy of BioJava jar file from BioJava website (www.biojava.org) 2. mysql-connector-java-3.0.10-stable-bin.jar an API for MySQL JDBC Connectivity (http://dev.mysql.com/downloads/connector/j/3.0.html) 3. xml-apis.jar, xercesImpl.jar XML APIs (http://xml.apache.org/xalan-j/downloads.html) 4. jakarta-oro-2.0.8.jar used by biojava (http://jakarta.apache.org/oro/) To get the latest versions of the JAR wp-content/uploads/2018/12 please visit aforementioned websites.