Severe acute respiratory syndrome coronavirus 2
aka: SARS-CoV-2
Coronavirus and the VBRC: a work in progress – email if you need help
The VBRC was developed for dsDNA viruses but has been adapted for coronaviruses.
- Only SARS-CoV-2 and closely related viruses will be added to this database.
- Other coronavirus genomes are available at NCBI or VIPRbrc.org
- VBRC tools can be used with FASTA and GenBank sequences.
- The VBRC provides unique tools that may be useful for the analysis of SARS-CoV-2
- see Tools menu for a list of software tools
- we also build software to tackle specific bioinformatics/virology problems, often in collaboration with virologists.
Coronavirus information
- SARS-CoV-2 genomics resources
- SARS-CoV-2 science news
- The disease: COVID-19
- General News – COVID-19
- Key scientific literature
- How to use SLACK
- Scientific Literature for UVic Res Group
- Acknowledgements
Analysis tools at the VBRC
We provide highly interactive software tools that enable users to view and analyze viral genomes. These tools can be accessed directly from the “Tools” drop-down menu on the top left of this page. These tools include:
- Viral Orthologous Clusters (VOCs) allows users to easily search the Coronaviridae database for protein sequences or genes of a known function or of a specific genome and then apply different analysis techniques to the search result. VOCs provides quick and easy access to tools like BLAST, multiple alignment, dotplots, and genome statistics.
- Base-By-Base is a whole genome pairwise and multiple alignment editor. The program highlights differences between pairs of alignments and allows the user to easily navigate large alignments of similar sequences. In addition to visualizing genomes and protein sequences, Base-By-Base allows the user to estimate simple phylogenetic trees, calculate the numbers of conserved and non-conserved sequence positions, and to perform a variety of comparative analyses through graphical interfaces.
- Viral Genome Organizer (VGO) allows users to browse genomes, viewing genes, ORFs, start/stop codons, nucleic acid composition, and orthologs when working with multiple genomes.
To start work: Check the Quick Start Tutorials (well worth 5 minutes) or just dive in by selecting a tool from the menu. Our usual starting place is the VOCs, which allows you to select data for use with the analysis tools.
Things you can do with VBRC tools:
- Compare SARS-CoV-19 genomes; edit alignment; display nucleotide differences.
 .
. - Select gene; select orthologs; show SNPs associated associated with sets of viruses.
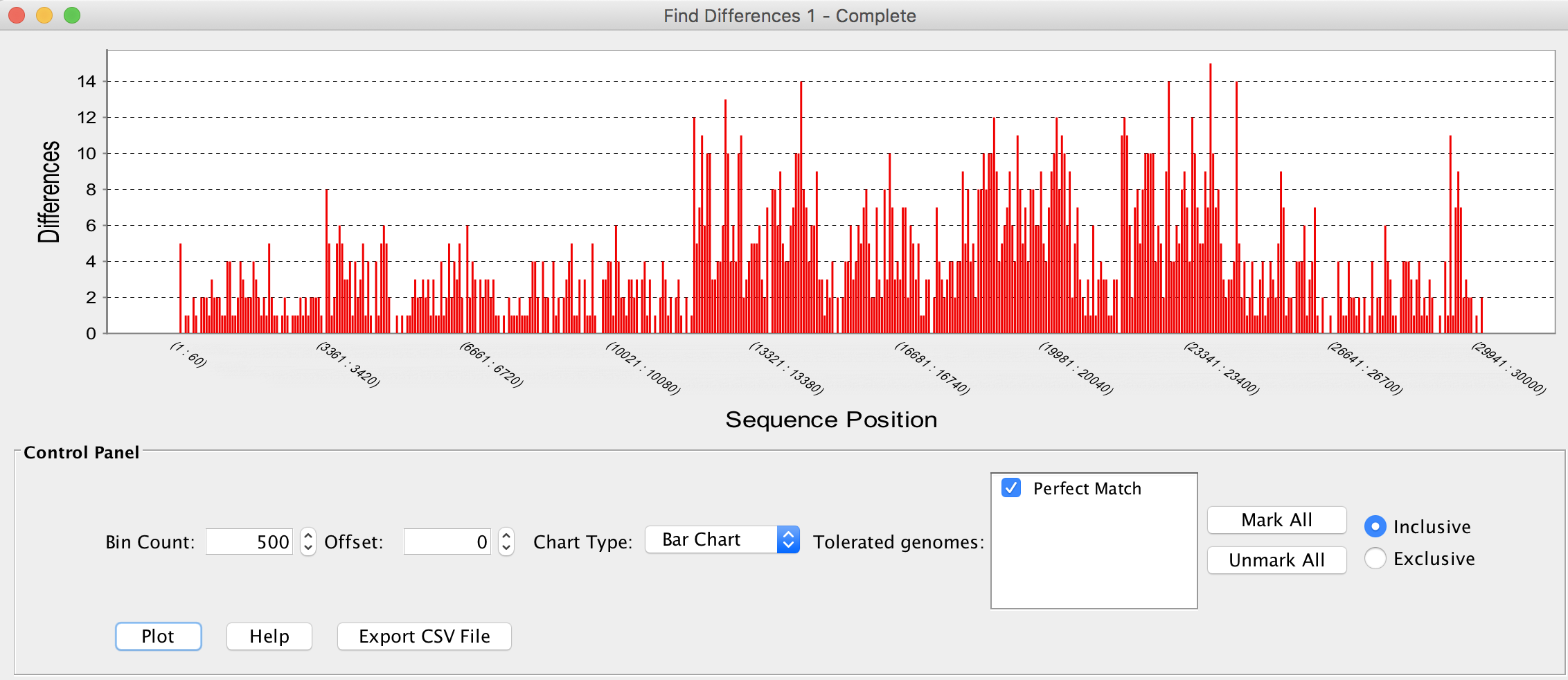 .
. - Align sets of proteins; edit alignment; highlight differences.
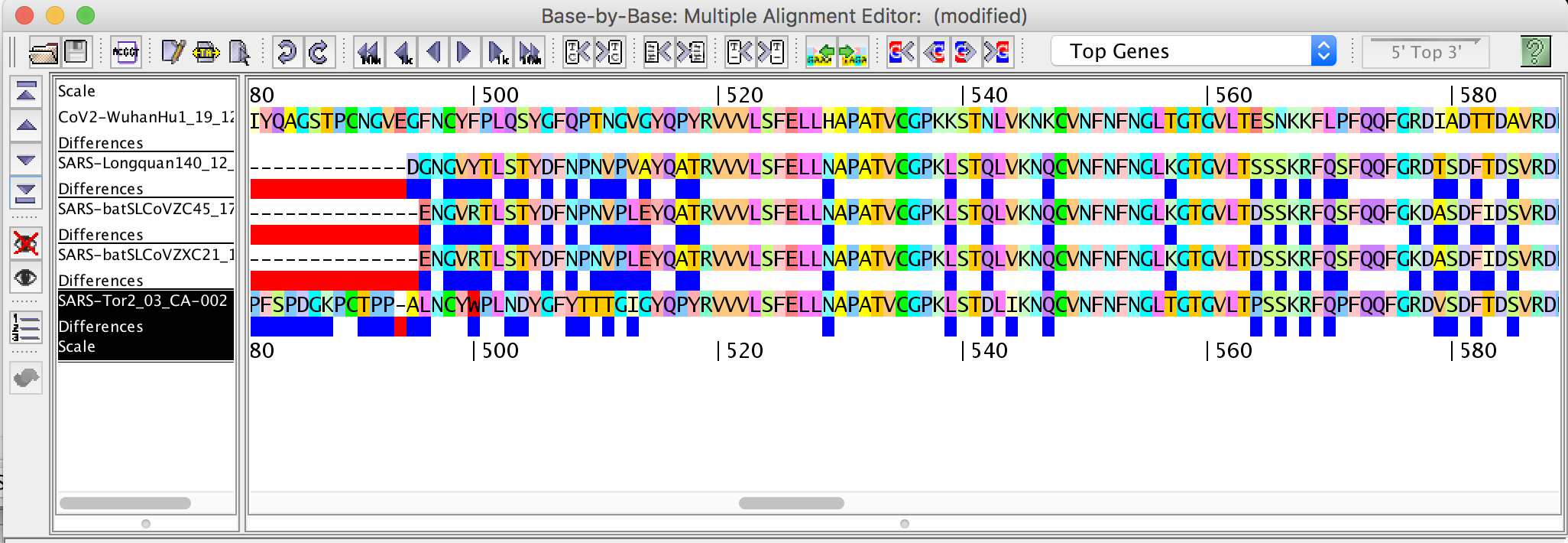 .
. - Annotate genomes with user comments; use as references in analyses.
Genomes excluded from VOCs because of errors
- Sequences with known errors will be listed here.
You can find the VBRC’s mandate here and more information on the home page.