j-CODEHOP PCR primer design
NOTE: This tool requires Java Version 8. Download it here: Java Runtime Environment.
j-CODEHOP is a tool integrated into Base-By-Base, and can be launched from the “Advanced” menu. The purpose of this tool is to design COnsensus DEgenerate Hybrid Oligonucleotide Primers (CODEHOPs) from amino acid sequence alignments.
Note: The first program created to make CODEHOPs was named CODEHOP (a Windows program), after which iCODEHOP (a web-based javascript program) was developed. Neither of these programs are available for use anymore, which is why j-CODEHOP (j standing for Java) was created. j-CODEHOP was modeled after CODEHOP and iCODEHOP, therefore literature about either of them may be used as
an alternative resource.
If you’re new to j-CODEHOP, click on the launch button (on the right) and use the j-CODEHOP Tutorial to learn the basic.
If you’re new to Base-By-Base, the basics are explained in the Quick Start Page, the How to doc and the Help Book.
If you would like more details on j-CODEHOP, please see the j-CODEHOP How-to Guide.
See the bottom of the page for further help.
j-CODEHOP can design PCR primers capable of amplifying distantly related genes. It has been used to identify and characterize new gene orthologs and paralogs in different plant, animal and bacterial species, as well as identify new pathogenic species. List of research papers that cite CODEHOP.
Note: a CODEHOP is a pool of related primers. Two CODEHOPs are required, acting as the 5′ and 3′ (forward and reverse) PCR primers.
j-CODEHOP is an interactive tool for designing CODEHOPs (primers) from conserved blocks of amino acids within aligned protein sequences. Each primer in the pool of CODEHOPs consists of both a 3′ degenerate core and a 5′ consensus sequence (also called the non-degenerate ‘clamp’ region) . Each 3′ degenerate core nucleotide sequence matches the nucleotides coding for the 3-4 highly conserved amino acids identified in the protein multiple alignment. The pool of primers represented within a CODEHOP differ in the 3′ degenerate core nucleotide sequence, providing all of the possible nucleotide sequence combinations that encode the same amino acid sequence. This provides a range of primers for the nucleotide coding region of the conserved amino acids, including previously unknown nucleic acid sequences. The 5′ consensus clamp sequence is created from a backtranslation of the 5-7 aa immediately 5’ of the degenerate core region. Since it is built from the most common nucleotide in each position of these codons, it increases the length of the primer without increasing degeneracy. This region is typically between 15 and 20 nucleotides, and its length can be adjusted by the user.
The 3’-degenerate structure of each CODEHOP pool allows the PCR amplification to have a broad specificity for distantly related target gene templates, while the 5’ consensus clamp allows for a robust amplification from PCR product templates during the later cycles of the amplification process.
Selection of the two optimal CODEHOPs is primarily based on minimal degeneracy across the 3’ degenerate cores and secondarily on the clamp scores, which indicates the quality of the matches between the 5’ clamp (a single sequence) and the amino acid sequence block given a codon usage table.
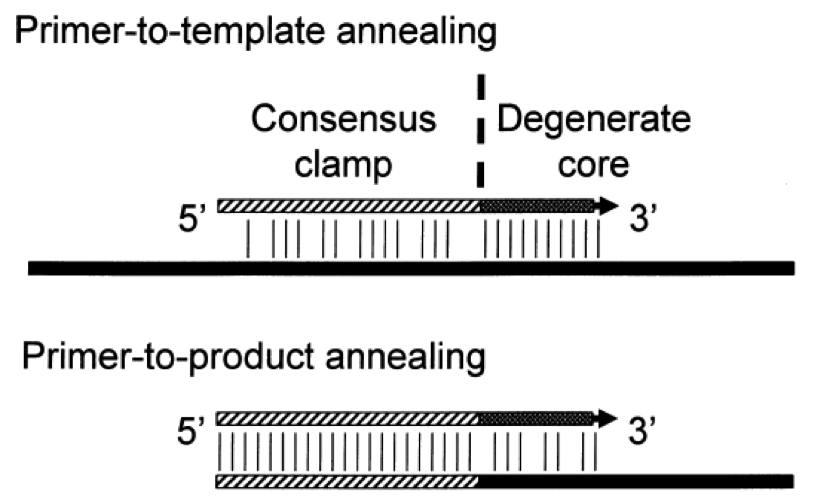
Figure 1. CODEHOP PCR primers target conserved motifs of 3–5 amino acids using a 3’ degenerate core containing all codon possibilities for the motif. The 5’ consensus clamp stabilizes the primer to the starting template. During subsequent rounds of amplification, the original primer sequence is a perfect match to the product. Annealing of the alternate primers from the degenerate primer pool to the PCR product is driven by the identical match with the 5’ consensus clamp; a consistent sequence on all primers. The primer-to-product annealing depicts the identical match between the 5’ consensus clamp and the product and the mismatches between the product and the variable degenerate region of the alternate primer. This strategy permits amplification of distantly related sequences with limited similarity to known family members.
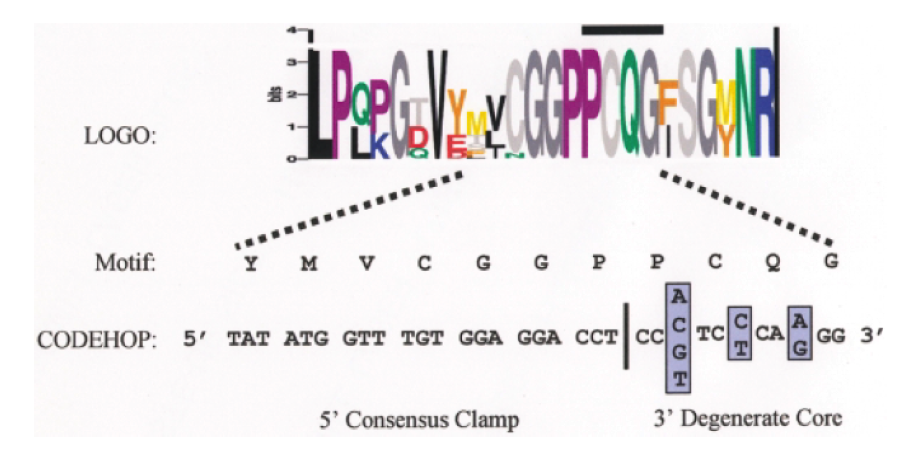
Figure 2. Anatomy of a CODEHOP PCR primer. A CODEHOP PCR primer is targeted to an amino acid sequence motif within a block of amino acids conserved between different divergent members of a protein family. The height of each amino acid is proportional to its degree of conservation. In this example the CODEHOP PCR primer is targeted to the highly conserved ‘PCQG’ motif and consists of a pool of 16 related primers in which the 3’ degenerate core contains all of the possible nucleotide sequences encoding the ‘PCQG’ motif. The 5’ consensus clamp, immediately upstream, contains the most probable nucleotide at each position flanking the ‘PCQG’ motif.
Using Base-by-Base
The Launch Tool under j-CODEHOP will automatically launch Base-By-Base. A FASTA file containing multiple amino acid sequences can be added under ‘File’, ‘Add Alignment/Sequence’.
If you are having any trouble with Base-By-Base, please see the “Getting Started Page” under Base-By-Base in the toolbar.
If you would like more details on j-CODEHOP, please see the j-CODEHOP How-to Guide.
j-CODEHOP can be used to predict PCR primers for amplification of distantly related gene sequences, from various families, genera, strains, etc.
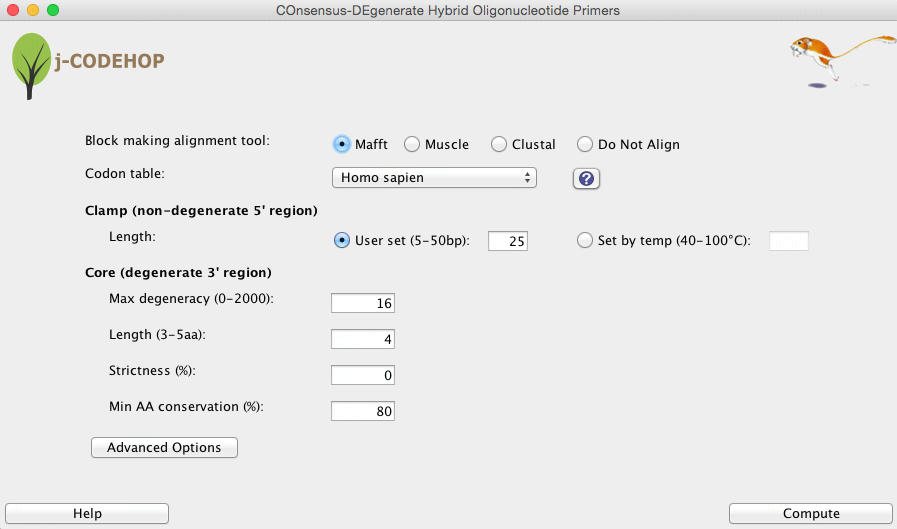
Figure 3. Initial window when starting j-CODEHOP displaying basic parameters for CODEHOP (primer) generation.
j-CODEHOP has been designed with an easy-to-use interface. Adjust your parameters, and click compute. The Help button includes ideas to generate more or less primers. A description of some parameters is given below. For a more in-depth description on all the j-CODEHOP parameters please see the j-CODEHOP How-to Guide.
Clamp Length can be set manually or determined by a annealing temperature calculation. Currently, the temperature calculation can only be performed when using a Mac OS X operating system. If “set by temperature” is selected, the clamp region is extended until the desired temperature is reached.
Max degeneracy measures how many different sequences are specified by the primer. The degeneracy of each position is the number of nucleotides appearing in it (1 to 4). The degeneracy of the primer is the product of the degeneracies of each position. Because the clamp region is a single consensus sequence, it does not add to the degeneracy within the CODEHOP pool.
Strictness is a percentage specifying a level of occurrence required for nucleotide inclusion. Strictness of 0 means that all nucleotides that actually appear in the position are included in the primer. Strictness of 1 means that only the most occurring nucleotide(s) in the position are included. Intermediate strictness values give behavior in between.
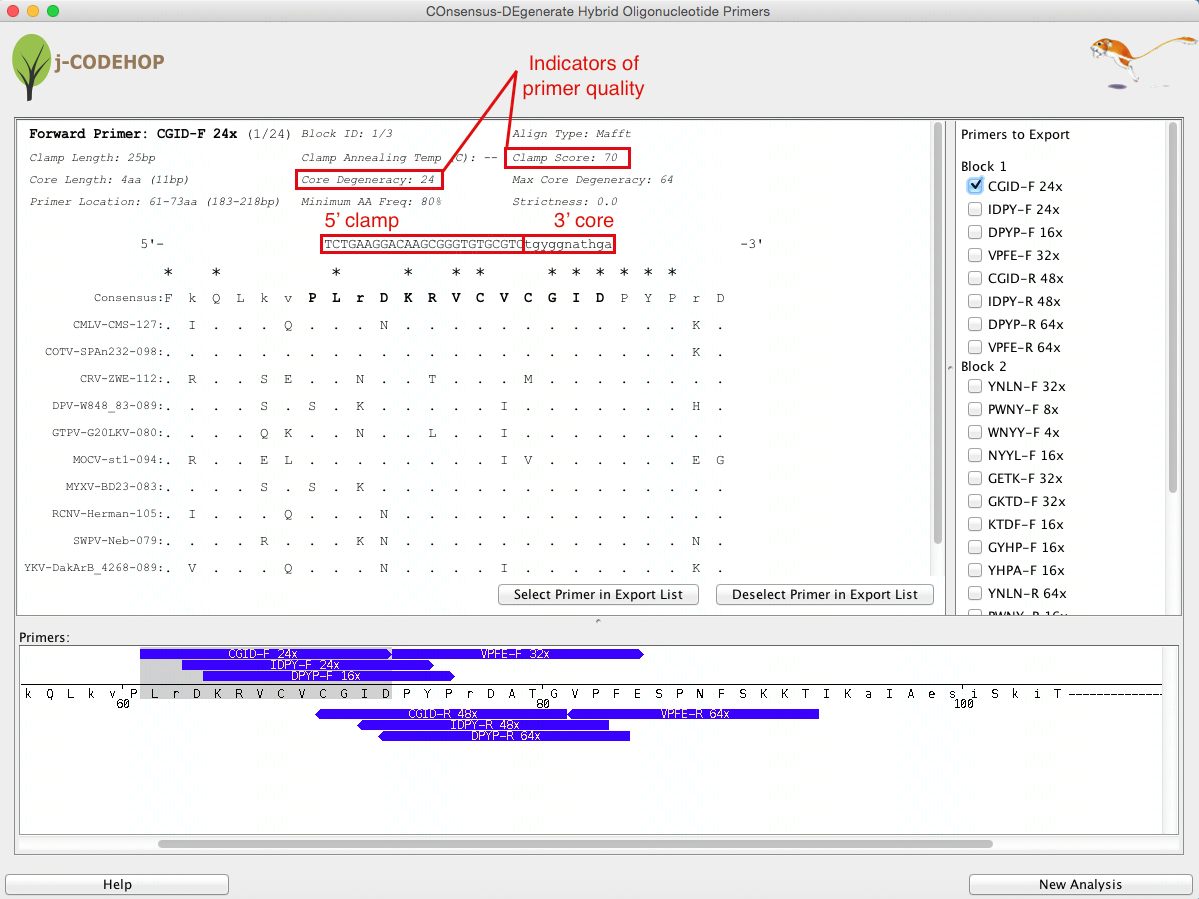
Figure 4. Window displaying specific information about one of the CODEHOPs generated, a small region of the primer location (to see the location of more primers the bottom window can slide across the sequence), as well as the list of all the CODEHOPs generated.
Clickable primers are displayed in the bottom panel and details of the selected primer are displayed in the top panel. An export feature is available to save primer information in a spreadsheet format that can be opened in a program such as Excel.

Figure 5. A forward and backward CODEHOP can be chosen form conserved blocks, with DNA between them, and can be used to study similar regions i other species to find unknown or distantly related genes.
To use the CODEHOP primers, choose a forward (5′) and backward (3′) primer from the list of generated primers that encompass the DNA segment to be amplified.
Getting Started
If you’re new to j-CODEHOP, click on the launch button (on the right) and use the j-CODEHOP Tutorial to learn the basics (or if you’re like us… just start clicking!).
The VBRC also provides additional help resources for j-CODEHOP:
- How to doc; short descriptions of certain analyses you might want to do.
- FAQs (Frequently Asked Questions)
- Finally, just email us a question and we’ll gladly help you out.
- The Viral Bioinformatics Resource software is published under the GNU General Public License version 3
References
If you use this resource please cite the relevant papers
Boyce R et al. (2009) iCODEHOP: a new interactive program for designing COnsensus-DEgenerate Hybrid Oligonucleotide Primers from multiply aligned protein sequences, Nucleic Acids Res. 37: 222-228.
Rose TM et al. (2003) CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design, Nucleic Acids Res. 31(13):3763-3766.
Rose TM et al. (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences, Nucleic Acids Res. 26(7):1628-1635.