Java Dot Plot Alignments
NOTE: This tool requires Java Version 8. Download it here: Java Runtime Environment.
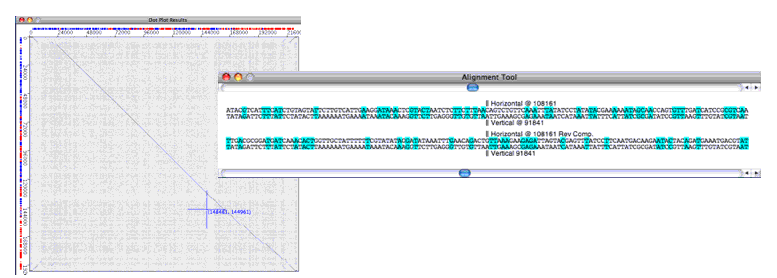
Java Dot Plot Alignments (JDotter) is a platform-independent Java interactive interface for the Linux version of Dotter, a widely used program for generating dotplots of large DNA or protein sequences. JDotter runs as a client-server application and can send new sequences to the Dotter program for alignment as well as rapidly access a repository of preprocessed dotplots. JDotter also interfaces with a sequence database or file system to display supplementary feature data. Thus, JDotter greatly simplifies access to dotplot data in laboratories that deal with large numbers of genomes and have a multi-platform organization.
With this program, you can:
- create whole-genome, sub-genome or protein dot plots from user-supplied sequences;
- quickly load pre-calculated plots of complete genomes from VOCs database;
- view and navigate the plots to pinpoint insertions/deletions and evaluate overall genome similarity;
- print/save dot plots in JDotter or JPEG format.
Getting Started
If you’re new to JDotter, click on the launch button (on the right) and use the Quick Start Page to learn the basics (or if you’re like us… just start clicking!).
The VBRC also provides additional help resources for JDotter:
- How to doc; short descriptions of certain analyses you might want to do.
- VBRC Help Book; our comprehensive reference manual.
- FAQs (Frequently Asked Questions)
- Finally, just email us a question and we’ll gladly help you out.
References
If you use this resource please cite the relevant papers (publication list).
Brodie, R., R. L. Roper, and C. Upton. 2004
JDotter: A Java Interface to Multiple DotPlots Generated by Dotter. Bioinformatics, 20, 279-281.
Papers which have cited JDotter
Troubleshooting
If your system does not launch Viral Orthologous Clusters, it is probably missing Java Runtime Environment 1.8. Please see Java Web Start Setup and Java Web Start Tips for help.