The Main Window
The main VOCs window is the user interface for searching the VOCs databases. It consists of three sections:
Within each of these 3 sections, you may use The Virus Selector
In the VOCs database, each gene (and it’s associated protein) is sorted into an ortholog group based on BLASTp scores. The main VOCs window allows you to search the database either by gene sequence, protein sequence or by ortholog group.
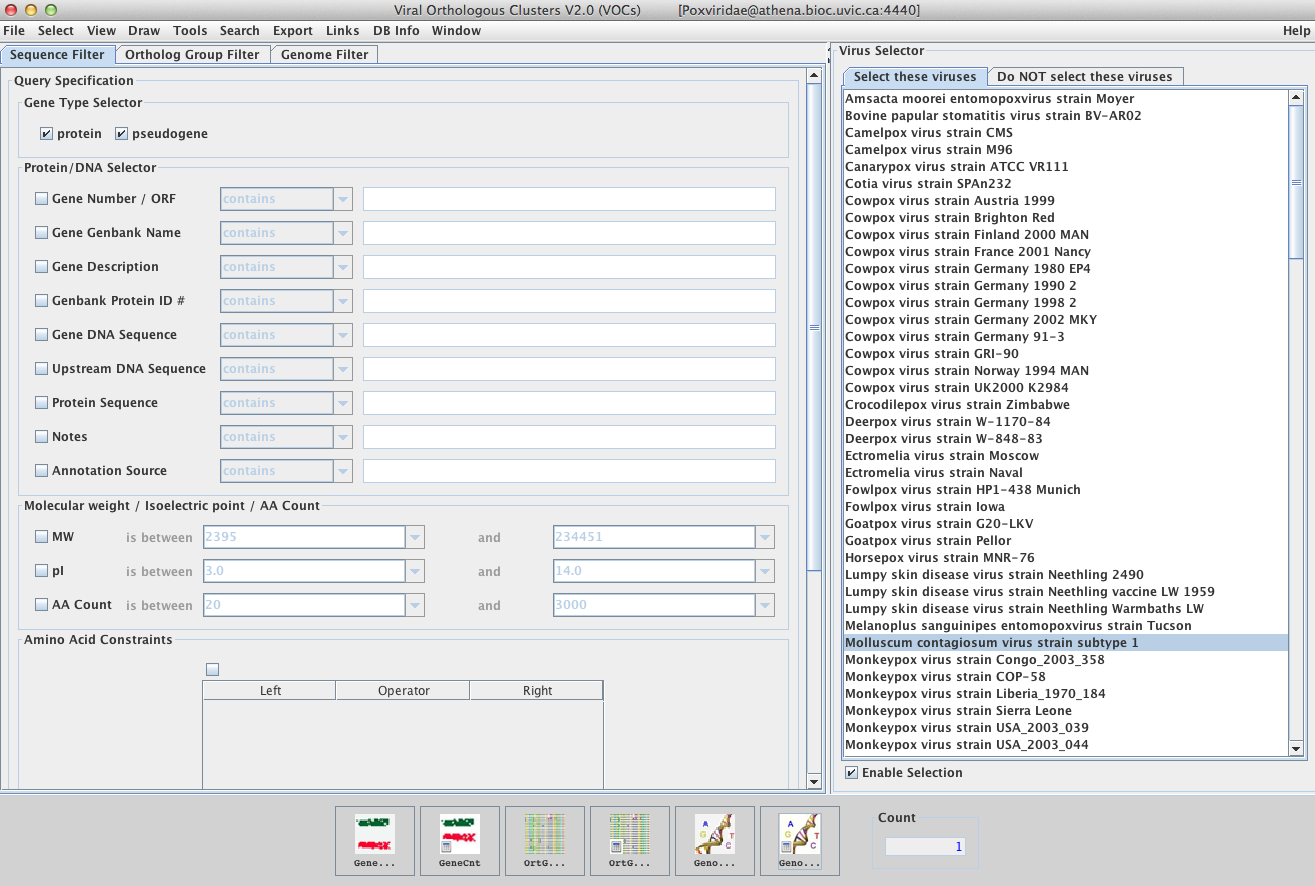
The Sequence Filter
Here you can enter search constraints and perform searches for the genes (or associated proteins) meeting your criteria. Possible constraints include gene type, gene name, DNA sequence, protein sequence, molecular weight, isoelectric point, amino acid count, % composition of amino acids, % composition of nucleotides etc.
The Ortholog Group Filter
In this part of VOCs you can enter search criteria specific to locating ortholog groups of interest. For example, you could potentially search for all ortholog groups containing genes of a specific genome.
The Genome Filter
Using a number of filtering parameters, this section of VOCs is used to refine the number of genomes listed in the 2 windows of the Virus Selector. This not only ensures that the list is less cumbersome to work with but also more specific to your needs.
The Virus Selector
Using this function, you may select one or more genomes from either of the two windows. You may choose to use only the genomes you select or alternatively, you may choose to exclude only the genomes you have selected. This is dependent on which of the windows you decide to activate (either “Select these viruses” or “Do NOT select these viruses”). To select multiple genomes use the ‘command’ or ‘ctrl’ button on your keyboard.
The Toolbar Buttons:
These buttons appear at the bottom of the main VOCs window.
Gene View (“GeneView”) displays the Gene Results Table listing the genes that fall within the constraints defined in the Sequence Filter, Ortholog Group Filter, Genome Filter and/or Virus Selector.
Gene Count (“GeneCnt”) counts the number of genes that fall within the constraints defined in the Sequence Filter, Ortholog Group Filter, Genome Filter and/or Virus Selector. The count will be displayed in the “Count” field located near the rightmost edge of the toolbar (you may need to expand the width of the main window if it is not visible).
Ortholog Group View (“OrtGrpView”) displays the Ortholog Groups table listing the ortholog groups that fall within the constraints defined in the Sequence Filter, Ortholog Group Filter, Genome Filter and/or Virus Selector.
Ortholog Group Count (“OrtGrpCnt”) counts the number of ortholog groups that fall within the constraints defined in the Sequence Filter, Ortholog Group Filter, Genome Filter and/or Virus Selector. The count will be displayed in the “Count” field located near the rightmost edge of the toolbar.
Genome View (“GenomeView”) displays the Genome Data table which lists all genomes that fall within the constraints defined in the Sequence Filter, Ortholog Group Filter, Genome Filter and/or Virus Selector.
Genome Count (“GenomeCnt”) counts the number of genomes that fall within the constraints defined in the Sequence Filter, Ortholog Group Filter, Genome Filter and/or Virus Selector. The count will be displayed in the “Count” field located near the rightmost edge of the toolbar.